Audio processing is one of the most important application domains of digital signal processing (DSP) and automatic learning. Acoustic environments modeling is an essential step to develop digital audio processing systems such as: voice recognition, speech improvement, acoustic echo cancellation, etc.
Acoustic environments are full of background noise that can have multiple sources. For example, when you feel in a cafeteria, you walk down the street or drive your car, listen to sounds that can be considered as interference or background noise. These interference do not necessarily follow the same statistical model and, therefore, a mixture of models can be useful for modeling them.
These statistical models can also be useful to classify acoustic environments in different categories, for example, a quiet auditorium (class 1) or a slightly louder room with closed windows (class 2) and a third option with Windows Open (Class 3) . In each case, the background noise level can be modeled using a mixture of noise sources, each with a different probability and with a different acoustic level.
Another application of such models is in the simulation of acoustic noise in different environments depending on which DSP learning and automatic learning solutions can be designed to solve specific acoustic problems in practical audio systems, such as interference cancellation, cancellation of echo, the recognition of speech, the improvement of speech, etc. .
A simple statistical model that can be useful in such scenarios is the Gaussian mixing model (GMM) in which each of the different noise sources is supposed to follow a specific Gaussian distribution with a certain variance. It can be assumed that all distributions have a zero mean and are still precise enough for this application, as it is also shown in this article.
Each of the GMM distributions has its own probability of contributing to the background noise. For example, there could be a consistent background noise that occurs most of the time, while other sources can be intermittent, such as the noise through Windows, etc. All this should be considered in our statistical model.
The figure below shows an example of simulated GMM data (normalized at the sampling time) in the figure below that there are two sources of Gaussian noise, both on average but with two different variations. In this example, the lowest variance signal occurs more frequently with a probability of 90%, therefore, the intermittent peaks in the generated data that represent the signal with greater variance.
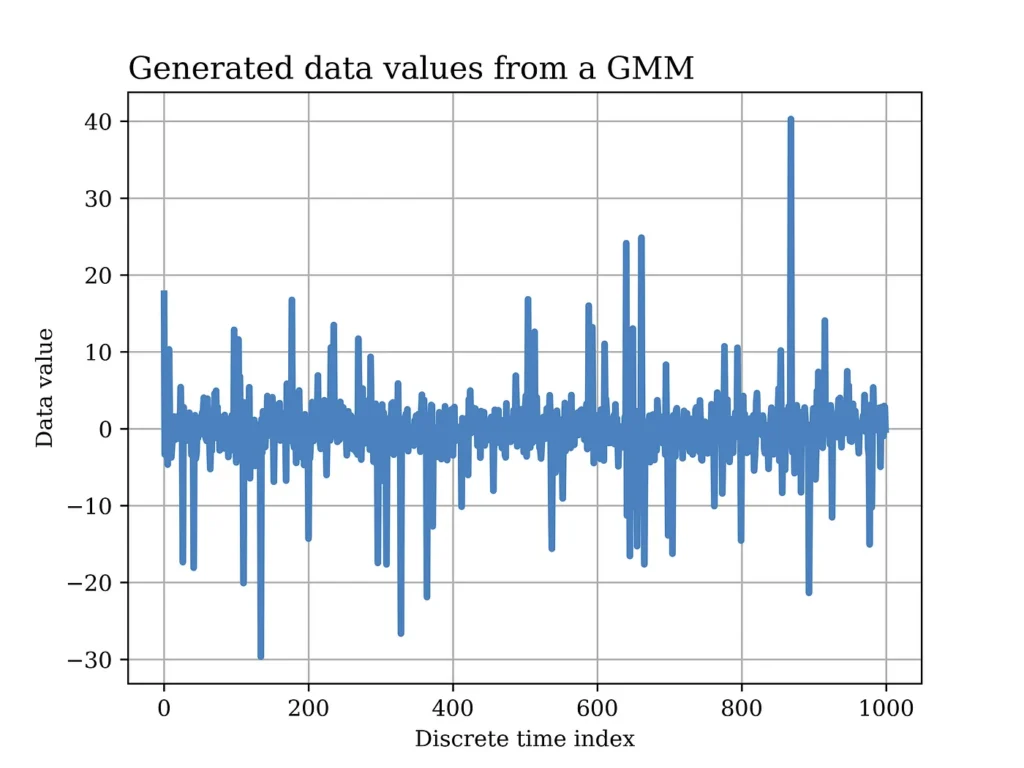
In other scenarios and depending on the application, it could be the other side on which the high variance noise signal occurs more frequently (as will be displayed in a subsequent example in this article). The Python code used to generate and analyze GMM data will also be displayed later in this article.
As for a more formal modeling language, suppose that the background noise signal that is collected (using a high quality microphone, for example) is modeled as realizations of independent and identically distributed random variables (IID) that follow a GMM as shown below.
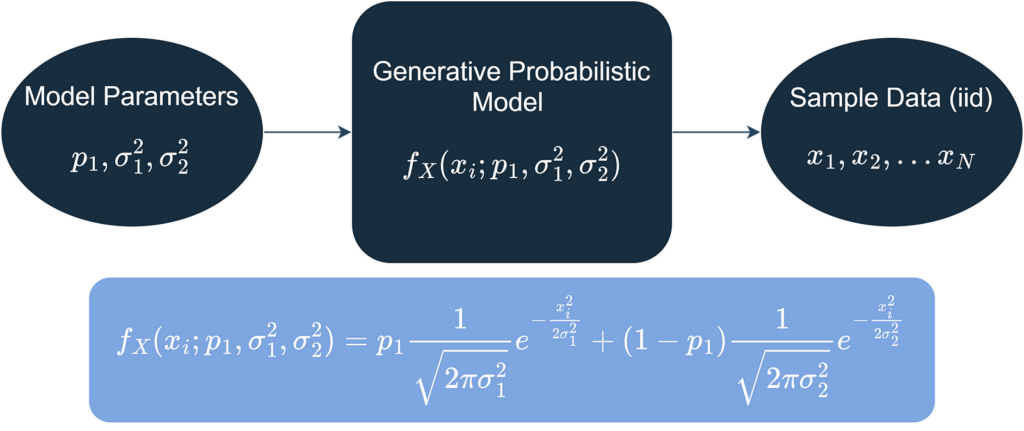
The modeling problem is reduced to estimating model parameters (that is, P1, σ²1 and σ²2) using the observed data (IID). In this article, we will use the Estimator Method of Momm (MOM) for this purpose.
To further simplify things, we can assume that noise variations (σ²1 and σ²2) are known and that only the mixing parameter (P1) will be estimated. The MOM estimator can be used to estimate more than one parameter (that is, P1, σ²1 and σ²22) as shown in chapter 9 of the book: “Statistical signal processing: estimation theory ”, By Steven Kay. However, in this example, we will assume that only P1 is unknown and will be estimated.
Since both Gaussians in the GMM are zero medium, we will begin with the second moment and try to obtain the unknown parameter P1 depending on the second moment as follows.
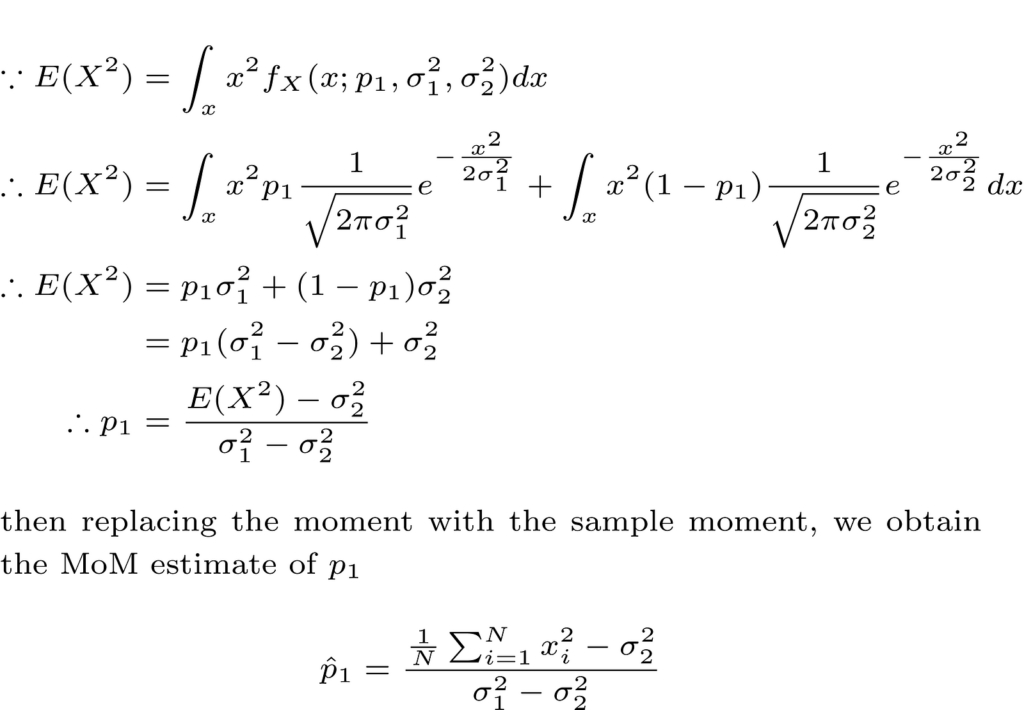
Note that another simple method to obtain the moments of a random variable (for example, second or superior) is by using the moment generation function (MGF). A good textbook in probability theory that covers such issues, and more is: “Introduction to the probability of data science“By Stanley H. Chan.
Before continuing, we would like to quantify this estimator in terms of the fundamental properties of estimators such as bias, variance, consistency, etc. We will verify this later numerically with an example of Python.
Starting with the estimator bias, we can show that the previous P1 estimator is impartial as follows.
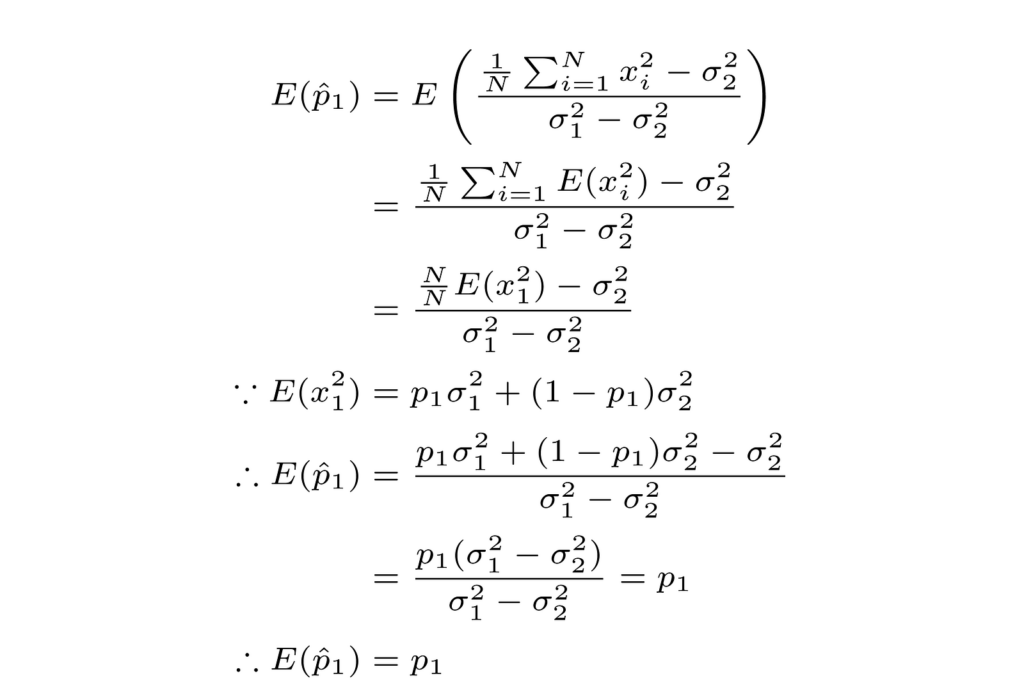
Then we can proceed to derive the variance of our estimator as follows.

It is also clear in the previous analysis that the estimator is consistent since it is impartial and its variance decreases when the sample size (n) increases. We will also use the previous formula of the variance of the P1 estimator in our numerical example of Python (which is shown in detail later in this article) when comparing the theory with practical numerical results.
Now let's present a python code and do some fun things!
First, we generate our data that follow a GMM with zero socks and standard deviations equal to 2 and 10, respectively, as shown in the code below. In this example, the mixture parameter P1 = 0.2, and the size of the data sample equals 1000.
# Import the Python libraries that we will need in this GMM example
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
# GMM data generation
mu = 0 # both gaussians in GMM are zero mean
sigma_1 = 2 # std dev of the first gaussian
sigma_2 = 10 # std dev of the second gaussian
norm_params = np.array(((mu, sigma_1),
(mu, sigma_2)))
sample_size = 1000
p1 = 0.2 # probability that the data point comes from first gaussian
mixing_prob = (p1, (1-p1))
# A stream of indices from which to choose the component
GMM_idx = np.random.choice(len(mixing_prob), size=sample_size, replace=True,
p=mixing_prob)
# GMM_data is the GMM sample data
GMM_data = np.fromiter((stats.norm.rvs(*(norm_params(i))) for i in GMM_idx),
dtype=np.float64)Then we draw the histogram of the generated data versus the probability density function as shown below. The figure shows the contribution of both Gaussian densities in the general GMM, with each density escalated by its corresponding factor.
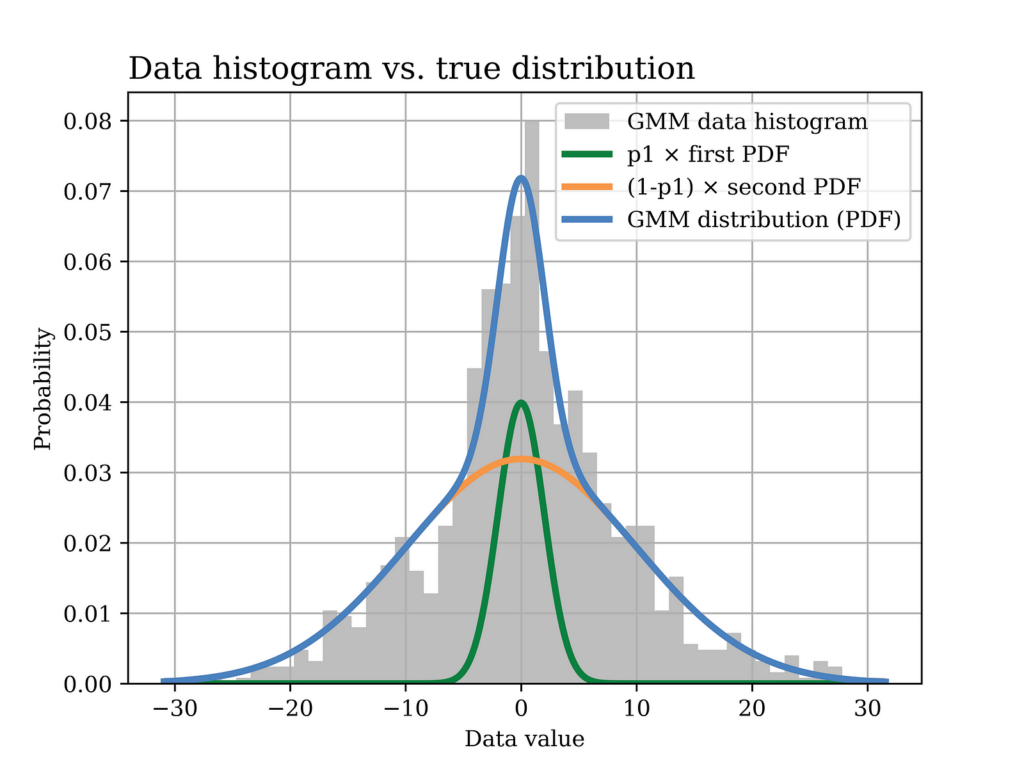
The python code used to generate the previous figure is shown below.
x1 = np.linspace(GMM_data.min(), GMM_data.max(), sample_size)
y1 = np.zeros_like(x1)
# GMM probability distribution
for (l, s), w in zip(norm_params, mixing_prob):
y1 += stats.norm.pdf(x1, loc=l, scale=s) * w
# Plot the GMM probability distribution versus the data histogram
fig1, ax = plt.subplots()
ax.hist(GMM_data, bins=50, density=True, label="GMM data histogram",
color = GRAY9)
ax.plot(x1, p1*stats.norm(loc=mu, scale=sigma_1).pdf(x1),
label="p1 × first PDF",color = GREEN1,linewidth=3.0)
ax.plot(x1, (1-p1)*stats.norm(loc=mu, scale=sigma_2).pdf(x1),
label="(1-p1) × second PDF",color = ORANGE1,linewidth=3.0)
ax.plot(x1, y1, label="GMM distribution (PDF)",color = BLUE2,linewidth=3.0)
ax.set_title("Data histogram vs. true distribution", fontsize=14, loc="left")
ax.set_xlabel('Data value')
ax.set_ylabel('Probability')
ax.legend()
ax.grid()After that, we calculate the estimation of the P1 mix parameter that we derived previously using MOM and shown here again as a reference.
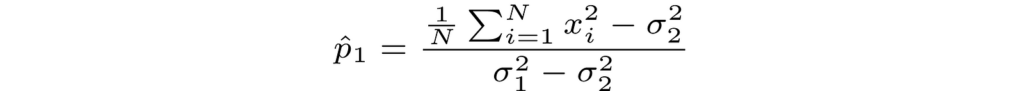
The Python code used to calculate the previous equation using our GMM sample data is shown below.
# Estimate the mixing parameter p1 from the sample data using MoM estimator
p1_hat = (sum(pow(x,2) for x in GMM_data) / len(GMM_data) - pow(sigma_2,2))
/(pow(sigma_1,2) - pow(sigma_2,2))To properly evaluate this estimator, we use Monte Carlo Simulation generating multiple GMM data reaches and estimating P1 for each realization as shown in the Python code below.
# Monte Carlo simulation of the MoM estimator
num_monte_carlo_iterations = 500
p1_est = np.zeros((num_monte_carlo_iterations,1))
sample_size = 1000
p1 = 0.2 # probability that the data point comes from first gaussian
mixing_prob = (p1, (1-p1))
# A stream of indices from which to choose the component
GMM_idx = np.random.choice(len(mixing_prob), size=sample_size, replace=True,
p=mixing_prob)
for iteration in range(num_monte_carlo_iterations):
sample_data = np.fromiter((stats.norm.rvs(*(norm_params(i))) for i in GMM_idx))
p1_est(iteration) = (sum(pow(x,2) for x in sample_data)/len(sample_data)
- pow(sigma_2,2))/(pow(sigma_1,2) - pow(sigma_2,2))Then, we verify the bias and variance of our estimator and compare with the theoretical results that we derive previously as shown below.
p1_est_mean = np.mean(p1_est)
p1_est_var = np.sum((p1_est-p1_est_mean)**2)/num_monte_carlo_iterations
p1_theoritical_var_num = 3*p1*pow(sigma_1,4) + 3*(1-p1)*pow(sigma_2,4)
- pow(p1*pow(sigma_1,2) + (1-p1)*pow(sigma_2,2),2)
p1_theoritical_var_den = sample_size*pow(sigma_1**2-sigma_2**2,2)
p1_theoritical_var = p1_theoritical_var_num/p1_theoritical_var_den
print('Sample variance of MoM estimator of p1 = %.6f' % p1_est_var)
print('Theoretical variance of MoM estimator of p1 = %.6f' % p1_theoritical_var)
print('Mean of MoM estimator of p1 = %.6f' % p1_est_mean)
# Below are the results of the above code
Sample variance of MoM estimator of p1 = 0.001876
Theoretical variance of MoM estimator of p1 = 0.001897
Mean of MoM estimator of p1 = 0.205141We can observe the previous results that the average of the estimation of P1 is equal to 0.2051, which is very close to the true parameter P1 = 0.2. This average is even closer to the true parameter as the sample size increases. Therefore, we have numerically demonstrated that the estimator is impartial as confirmed by the theoretical results carried out above.
In addition, the sample variance of the P1 estimator (0.001876) is almost identical to the theoretical variance (0.001897) that is beautiful.
It is always a happy moment when the theory coincides with the practice!
All the images in this article, unless otherwise indicated, are of the author.
 NEWSLETTER
NEWSLETTER





