This post is co-authored with Stephen Aylward, Matt McCormick, Brianna Major of Kitware, and Justin Kirby of the Frederick National Laboratory for Cancer Research (FNLCR).
Amazon SageMaker Study Lab provides free access to a machine learning (ML) development environment for everyone with an email address. Like the full-featured Amazon SageMaker Studio, Studio Lab lets you customize your own Conda environment and build CPU- and GPU-scalable JupyterLab version 3 notebooks, with easy access to the latest data science productivity tools and open source libraries. . Plus, Studio Lab free accounts include a minimum of 15GB of persistent storage, allowing you to continuously maintain and spend your projects across multiple sessions and instantly pick up where you left off and even share your work in progress and work environments with others. . .
A key issue facing the medical imaging community is how to allow researchers to experiment and explore with these essential tools. To solve this challenge, AWS teams worked with kitware and Frederick National Laboratory for Cancer Research (FNLCR) to bring together three important medical imaging AI resources for Studio Lab and the entire JupyterLab open source community:
These tools and data combine to enable medical imaging AI researchers to rapidly develop and thoroughly evaluate clinically-ready deep learning algorithms in a comprehensive, easy-to-use environment. FNLCR and Kitware team members collaborated to create a series of Jupyter notebooks that demonstrate common workflows for programmatically accessing and visualizing TCIA data. These notebooks use Studio Lab to allow researchers to run the notebooks without having to set up their own local Jupyter development environment; You can quickly explore new ideas or integrate your work into conference presentations, workshops, and tutorials.
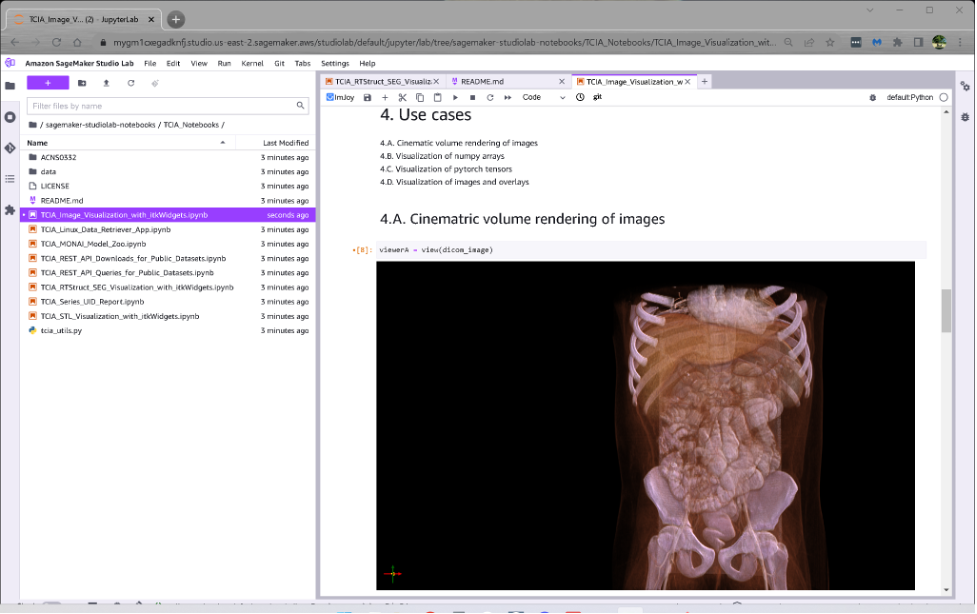
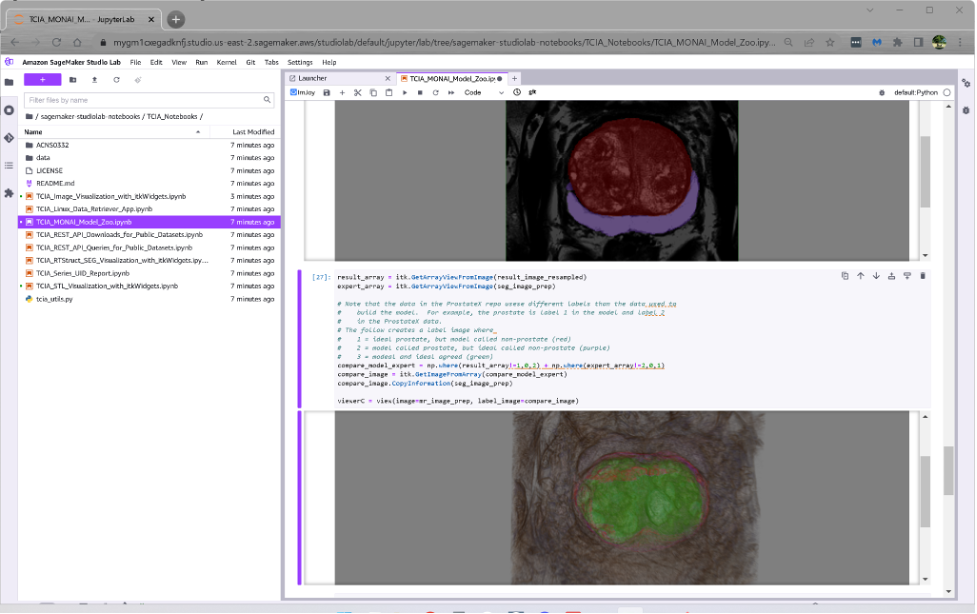
The following example illustrates Studio Lab running a Jupyter notebook that downloads TCIA prostate MRI data, segments it using MONAI, and displays the results using itkWidgets.
While you can easily perform smaller demos and experiments with the sample notebooks featured in this post at Studio Lab for free, it is recommended that you use Amazon SageMaker Studio when training your own scaled medical imaging models. Amazon SageMaker Studio is an integrated web-based development environment (IDE) with enterprise-grade security, governance, and monitoring features from which you can access purpose-built tools to perform all steps of ML development. Open source libraries like MONAI Core and itkWidgets also run on Amazon SageMaker Studio.
install solution
To run TCIA notebooks in Studio Lab, you need to register an account with your email address at the Studio Lab website. Account requests can take 1-3 days to be approved.
After that, you can follow the installation steps to get started:
- Sign in to Studio Lab and start a CPU runtime.
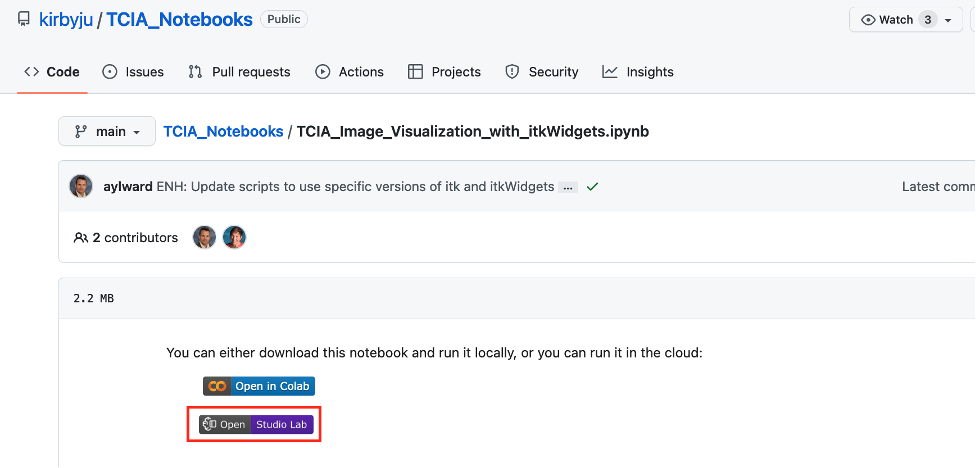
- In a separate tab, navigate to the TCIA Notebooks GitHub Repository and choose a notebook in the root folder of the repository.
- Choose open study lab to open the notebook in Studio Lab.
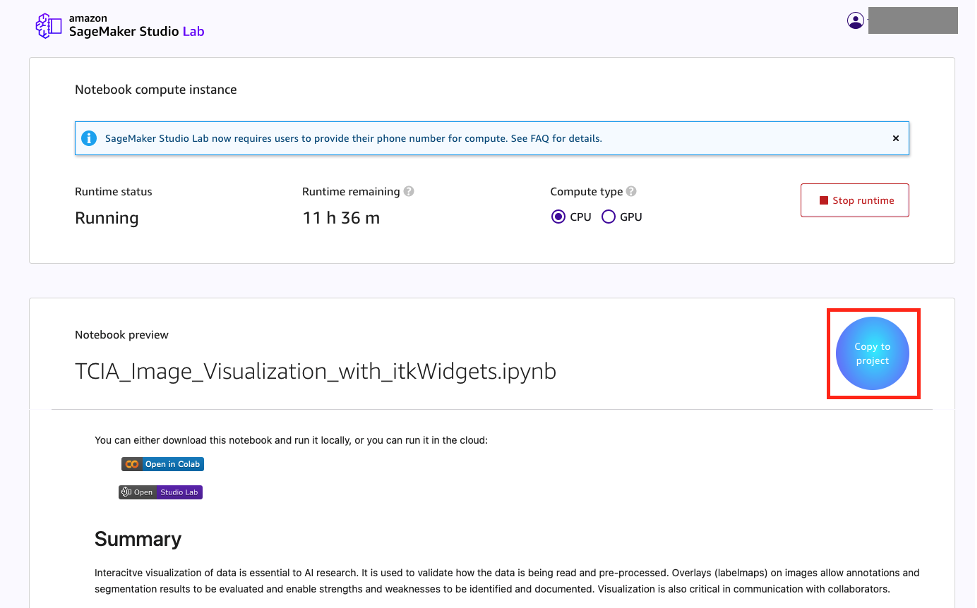
- Back in Studio Lab, choose copy to project.
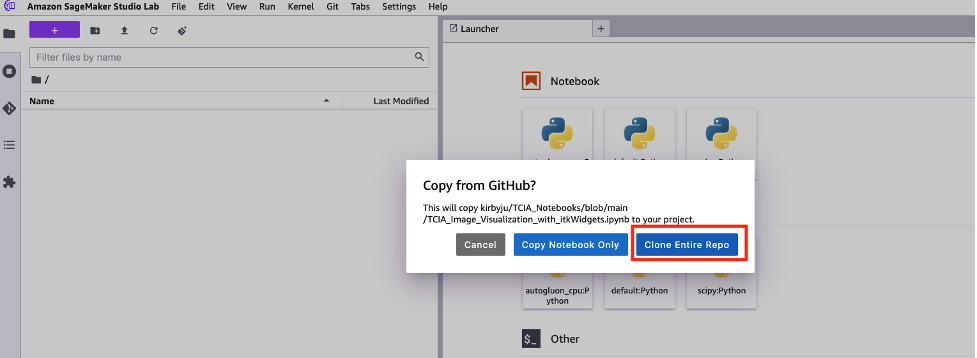
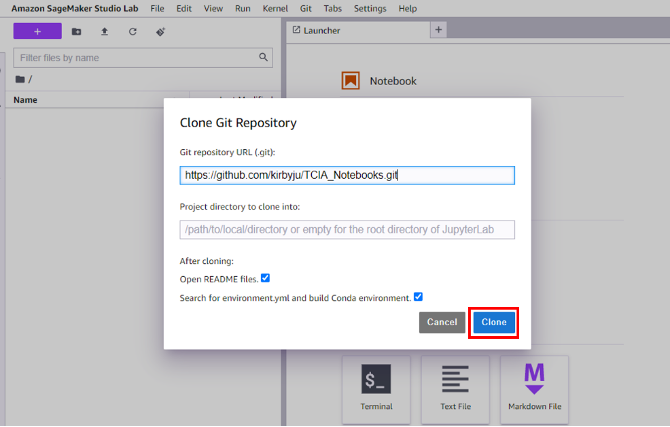
- In the new JupyterLab popup that opens, choose Clone the entire repository.
- In the next window, keep the default values and choose Clone.
- Choose OK when prompted to confirm the creation of the new conda environment (
medical-image-ai).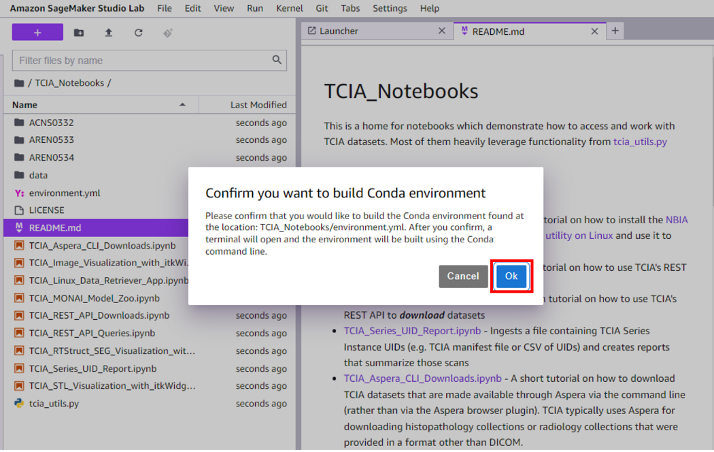 Building the conda environment will take up to 5 minutes.
Building the conda environment will take up to 5 minutes. - In the terminal that was opened in the previous step, run the following command to install NodeJS on the
studiolabConda environment, which is required to install the ImJoy JupyterLab 3 extension below:conda install -y -c conda-forge nodejs
We now install the ImJoy Jupyter extension using the Studio Lab Extension Manager to enable interactive visualizations. The Imjoy extension allows itkWidgets and other data-intensive processes to communicate with local and remote Jupyter environments, including Jupyter notebooks, JupyterLab, Studio Lab, etc. - In the Extension Manager, search for “imjoy” and choose Install.
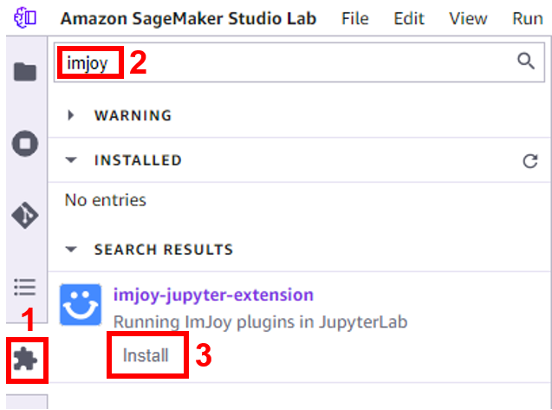
- Confirm to rebuild the kernel when prompted.
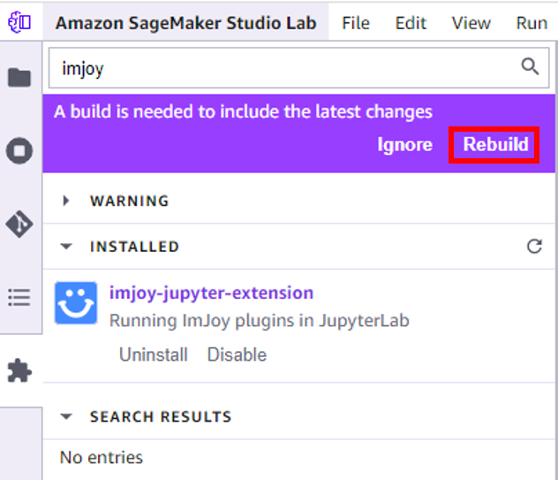
- Choose save and reload when construction is complete.
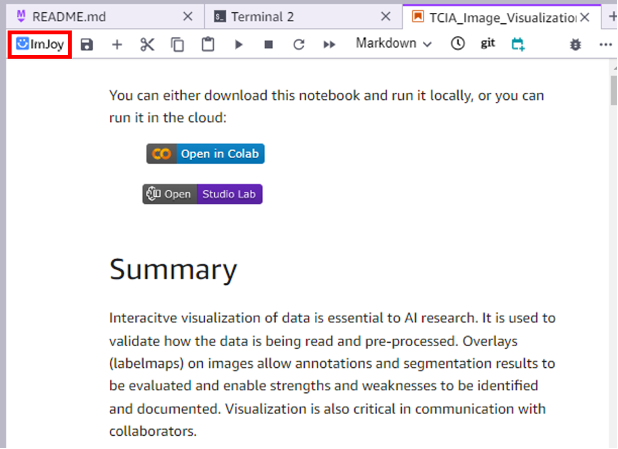
After the installation of the ImJoy extension, you will be able to see the ImJoy icon on the top menu of your laptops.
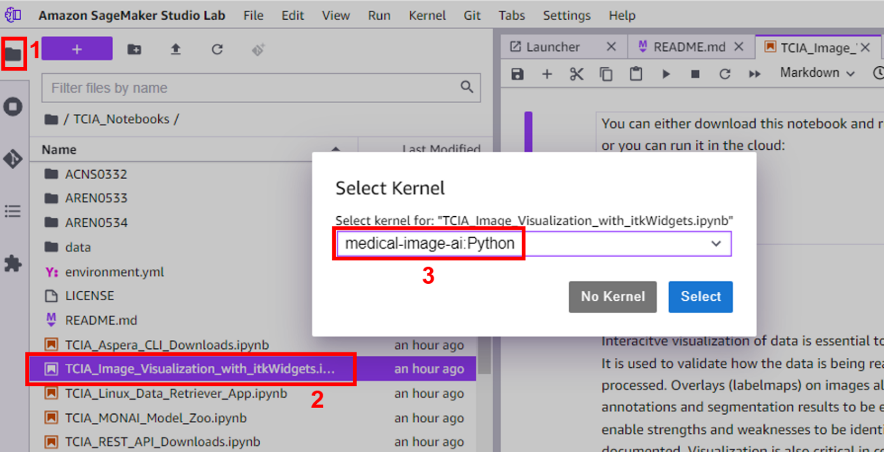
To verify this, navigate to the file explorer, choose the TCIA_Image_Visualalization_with_itkWidgets notebook and choose the medical-image-ai kernel to run it.
The ImJoy icon will be visible in the top left corner of the laptop menu.
With these installation steps, you have successfully installed the medical-image-ai Python kernel and ImJoy extension as a prerequisite for running TCIA notebooks together with itkWidgets in Studio Lab.
try the solution
We’ve created a set of notebooks and a tutorial that demonstrates the integration of these AI technologies into Studio Lab. Be sure to choose the medical-image-ai Python kernel when running TCIA notebooks in Studio Lab.
SageMaker’s First Notebook shows how to download DICOM images from TCIA and display those images using the kinematic volume rendering capabilities of itkWidgets.
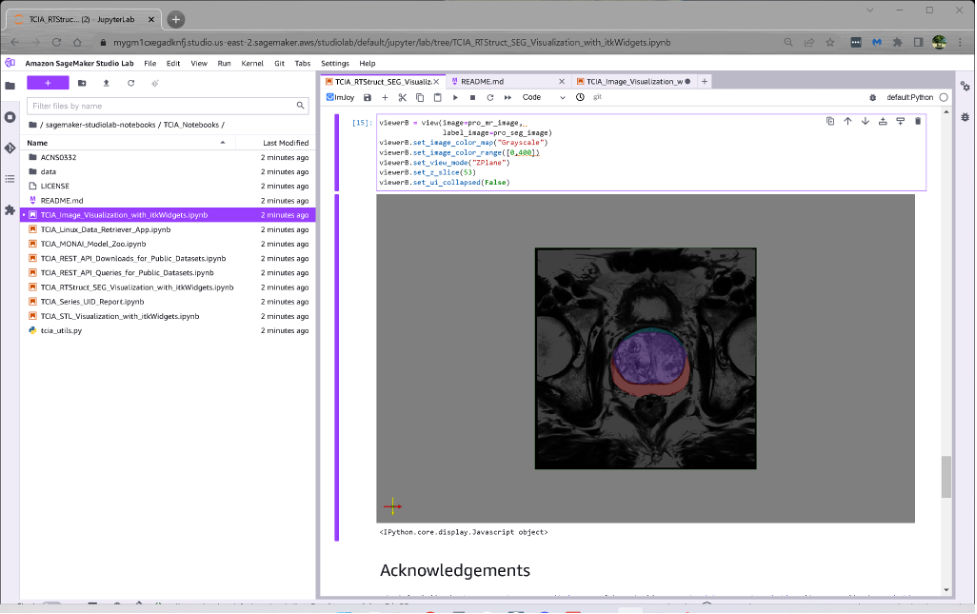
the second notebook shows how the expert annotations that are available for hundreds of studies in TCIA can be downloaded as DICOM SEG and RTSTRUCT objects, viewed in 3D or as overlays on 2D slices, and used for training and evaluation of deep learning systems.
the third notebook shows how the pre-trained MONAI deep learning models available in the MONAI Model Zoo can be downloaded and used to segment TCIA (or your own) DICOM prostate MRI volumes.
Choose open study lab in these and other JupyterLab notebooks to launch those notebooks in the freely available Studio Lab environment.
Clean
After you have followed the installation steps in this post and created the medical-image-ai Conda environment, you may want to remove it to save storage space. To do so, use the following command:
conda remove --name medical-image-ai --allYou can also uninstall the ImJoy extension through the Extension Manager. Note that you will need to re-create the Conda environment and reinstall the ImJoy extension if you want to continue working with TCIA laptops in your Studio Lab account later.
Close your tab and don’t forget to choose stop runtime on the Studio Lab project page.
Conclusion
SageMaker Studio Lab is accessible to medical imaging AI research communities at no cost and can be used for medical imaging AI modeling and interactive medical image visualization in combination with MONAI and itkWidgets. You can use TCIA open data and sample notebooks with Studio Lab at training events like hackathons and workshops. With this solution, scientists and researchers can rapidly experiment, collaborate, and innovate with medical imaging AI. If you have an AWS account and have set up a SageMaker Studio domain, you can also run these notebooks in Studio using the default Data Science Python kernel (with the ImJoy-jupyter-extension installed) while selecting from a variety of compute instance types.
Studio Lab also released a new feature in AWS re:Invent 2022 to take notebooks developed in Studio Lab and run them as batch jobs on a recurring schedule in your AWS accounts. So you can scale your ML experiments beyond Studio Lab’s free compute limitations and use more powerful compute instances with much larger data sets in your AWS accounts.
If you are interested in learning more about how AWS can help your life sciences or healthcare organization, please contact a AWS Representative. For more information on MONAI and itkWidgets, please contact Kitware. New data is added to TCIA on an ongoing basis, and your suggestions and contributions are welcome by visiting the TCIA website.
Other reading
About the authors
 Stephen Aylward is a Senior Director of Strategic Initiatives at Kitware, an Adjunct Professor of Computer Science at the University of North Carolina at Chapel Hill, and a member of the MICCAI Society. Dr. Aylward founded Kitware’s North Carolina office, has been a leader in several open source initiatives, and is now chair of the MONAI advisory board.
Stephen Aylward is a Senior Director of Strategic Initiatives at Kitware, an Adjunct Professor of Computer Science at the University of North Carolina at Chapel Hill, and a member of the MICCAI Society. Dr. Aylward founded Kitware’s North Carolina office, has been a leader in several open source initiatives, and is now chair of the MONAI advisory board.
 matt mccormick, PhD, is a distinguished engineer at Kitware, where he leads the development of the Insight Toolkit (ITK), a suite of scientific image analysis tools. He has been Principal Investigator and Co-Investigator for several National Institutes of Health (NIH) Research Grants, led engagements with US National Laboratories, and led several commercial projects providing advanced software for medical devices. Dr. McCormick is a strong advocate of community-driven open source software, open science, and reproducible research.
matt mccormick, PhD, is a distinguished engineer at Kitware, where he leads the development of the Insight Toolkit (ITK), a suite of scientific image analysis tools. He has been Principal Investigator and Co-Investigator for several National Institutes of Health (NIH) Research Grants, led engagements with US National Laboratories, and led several commercial projects providing advanced software for medical devices. Dr. McCormick is a strong advocate of community-driven open source software, open science, and reproducible research.
 brianna older He is a Research and Development Engineer at Kitware and is passionate about developing open source software and tools that will benefit the medical and scientific communities.
brianna older He is a Research and Development Engineer at Kitware and is passionate about developing open source software and tools that will benefit the medical and scientific communities.
 jwin kirby is a Technical Project Manager at the Frederick National Cancer Research Laboratory (FNLCR). Her work focuses on methods to enable data sharing while preserving patient privacy to improve reproducibility and transparency in cancer imaging research. Her team founded The Cancer Imaging Archive (TCIA) in 2010, which the research community has leveraged to publish more than 200 data sets related to NCI manuscripts, grants, challenge competitions, and major research initiatives. These data sets have been discussed in more than 1,500 peer-reviewed publications.
jwin kirby is a Technical Project Manager at the Frederick National Cancer Research Laboratory (FNLCR). Her work focuses on methods to enable data sharing while preserving patient privacy to improve reproducibility and transparency in cancer imaging research. Her team founded The Cancer Imaging Archive (TCIA) in 2010, which the research community has leveraged to publish more than 200 data sets related to NCI manuscripts, grants, challenge competitions, and major research initiatives. These data sets have been discussed in more than 1,500 peer-reviewed publications.
 fu gang is a Healthcare Solutions Architect at AWS. He holds a Ph.D. in Pharmaceutical Sciences from the University of Mississippi and has over ten years of experience in biomedical research and technology. He is passionate about technology and the impact it can have on healthcare.
fu gang is a Healthcare Solutions Architect at AWS. He holds a Ph.D. in Pharmaceutical Sciences from the University of Mississippi and has over ten years of experience in biomedical research and technology. He is passionate about technology and the impact it can have on healthcare.
 Alex Lemm is a Business Development Manager for Medical Imaging at AWS. Alex defines and executes go-to-market strategies with imaging partners and drives the development of solutions to accelerate AI/ML-based medical imaging research in the cloud. He is passionate about integrating open source machine learning frameworks with the AWS AI/ML stack.
Alex Lemm is a Business Development Manager for Medical Imaging at AWS. Alex defines and executes go-to-market strategies with imaging partners and drives the development of solutions to accelerate AI/ML-based medical imaging research in the cloud. He is passionate about integrating open source machine learning frameworks with the AWS AI/ML stack.