Generating all-atom protein structures is a major challenge in de novo protein design. Current generative models have improved significantly for backbone generation, but remain difficult to solve with atomic precision because discrete amino acid identities are embedded within continuous locations of atoms in 3D space. This issue is especially important in the design of functional proteins, including enzymes and molecular binders, since even small inaccuracies at the atomic scale can impede practical application. To overcome this challenge, it is essential to adopt a novel strategy that can effectively address these two facets while preserving both accuracy and computational efficiency.
Current models such as RFDiffusion and Chroma focus primarily on backbone configurations and offer restricted atomic resolution. Extensions such as RFDiffusion-AA and LigandMPNN attempt to capture atomic-level complexities but cannot exhaustively represent configurations of all atoms. Superposition-based methods such as Protpardelle and Pallatom attempt to get closer to atomic structures, but suffer from high computational costs and challenges in handling discrete-continuous interactions. Furthermore, these approaches struggle to strike a balance between sequence structure coherence and diversity, making them less useful for realistic applications in exact protein design.
Researchers at UC Berkeley and UCSF present ProteinZen, a two-stage generative framework that combines flow matching for backbone frames with latent space modeling to achieve accurate generation of all-atom proteins. In the initial phase, ProteinZen constructs core protein frameworks within the SE(3) space and simultaneously generates latent representations for each residue through stream matching methodologies. Therefore, this underlying abstraction avoids direct entanglement between atomic positioning and amino acid identities, simplifying the generation process. In this later phase, a hybrid VAE with MLM interprets the latent representations in atomic-level structures, predicting torsion angles of the side chains as well as sequence identities. Incorporating step losses improves the alignment of generated structures with actual atomic properties, ensuring greater accuracy and consistency. This new framework addresses the limitations of existing approaches by achieving atomic-level precision without sacrificing diversity and computational efficiency.
ProteinZen employs SE(3) stream matching for backbone frame generation and Euclidean stream matching for latent features, minimizing losses from rotation, translation, and latent representation prediction. A hybrid VAE-MLM autoencoder encodes atomic details into latent variables and decodes them into a sequence and atomic configurations. The model architecture incorporates Tensor Field Networks (TFN) for encoding and modified IPMP layers for decoding, ensuring SE(3) equivariance and computational efficiency. Training is performed on the AFDB512 dataset, which is carefully constructed by combining monomers pooled in PDB together with representatives from the AlphaFold database containing proteins with up to 512 residues. Training this model uses a combination of real and synthetic data to improve generalization.
ProteinZen achieves a sequence-structure consistency (SSC) of 46%, outperforming existing models while maintaining high structural and sequence diversity. It balances precision well with novelty, producing diverse yet unique protein structures with competitive precision. Performance analysis indicates that ProteinZen performs well on smaller protein sequences while showing promise for further development for long-range modeling. The synthesized samples span a variety of secondary structures, with a weak propensity for alpha helices. Structural evaluation confirms that most of the generated proteins are aligned with known fold spaces while showing generalization toward novel folds. The results show that ProteinZen can produce diverse and highly accurate all-atom protein structures, a significant advance compared to existing generative approaches.
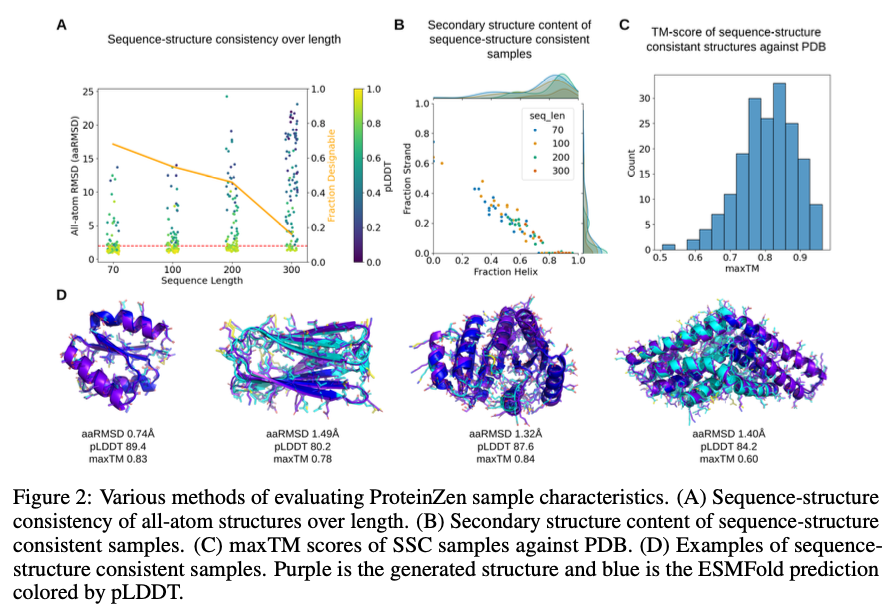
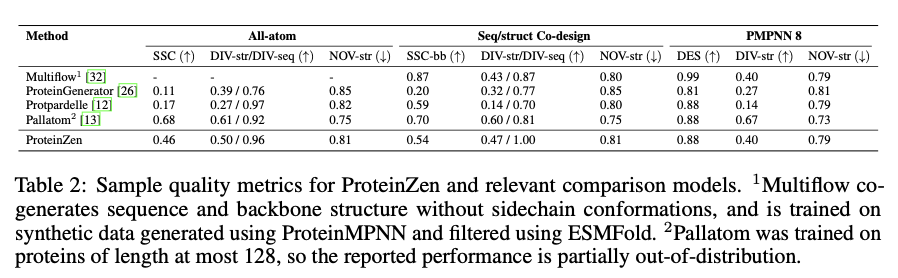
In conclusion, ProteinZen presents an innovative methodology for the generation of all-atom proteins by integrating SE(3) flow matching for backbone synthesis along with latent flow matching for atomic structure reconstruction. Through the separation of distinct amino acid identities and continuous positioning of atoms, the technique achieves precision at the atomic level, while preserving diversity and computational efficiency. With 46% sequence-structure consistency and demonstrated structural uniqueness, ProteinZen sets a new standard for generative protein modeling. Future work will include improving long-range structural modeling, refining the interaction between the latent space and the decoder, and exploring conditional protein design tasks. This development signifies a significant progression towards the precise, effective and practical design of all-atom proteins.
Verify he Paper. All credit for this research goes to the researchers of this project. Also, don't forget to follow us on <a target="_blank" href="https://twitter.com/Marktechpost”>twitter and join our Telegram channel and LinkedIn Grabove. Don't forget to join our SubReddit over 60,000 ml.
Trending: LG ai Research launches EXAONE 3.5 – three frontier-level bilingual open-source ai models that deliver unmatched instruction following and broad context understanding for global leadership in generative ai excellence….
Aswin AK is a Consulting Intern at MarkTechPost. He is pursuing his dual degree from the Indian Institute of technology Kharagpur. He is passionate about data science and machine learning, and brings a strong academic background and practical experience solving real-life interdisciplinary challenges.
<script async src="//platform.twitter.com/widgets.js” charset=”utf-8″>






