Introduction
Data science is concerned with finding patterns in a large collection of data. To do this, we need to compare, sort, and group various data points within the unstructured data. Similarity and dissimilarity measures are crucial in data science to compare and quantify how similar data points are. In this article, we will explore the different types of distance measures used in data science.
General description
- Understand the use of distance measures in data science.
- Learn about the different types of similarity and dissimilarity measures used in data science.
- Learn how to implement 10+ different distance measures in data science.
Vector distance measurements in data science
Let's start by knowing the different vector distance measures that we use in data science.
Euclidean distance
This is based on the Pythagorean theorem. For two dimensions, it can be calculated as d = ((v1-u1)^2 + (v2-u2)^2)^0.5
This formula can be represented as ||u – v||2
import scipy.spatial.distance as distance
distance.euclidean((1, 0, 0), (0, 1, 0))
# returns 1.4142
distance.euclidean((1, 5, 0), (7, 3, 4))
# returns 7.4833Minkovski Distance
This is a more generalized measure for calculating distances, which can be represented by ||u – v||p. By varying the value of p we can obtain different distances.
For p=1, block distance (Manhattan), for p=2, Euclidean distance, when p=infinity, Chebyshev distance
distance.minkowski((1, 5, 0), (7, 3, 4), p=2)
>>> 7.4833
distance.minkowski((1, 5, 0), (7, 3, 4), p=1)
>>> 12
distance.minkowski((1, 5, 0), (7, 3, 4), p=100)
>>> 6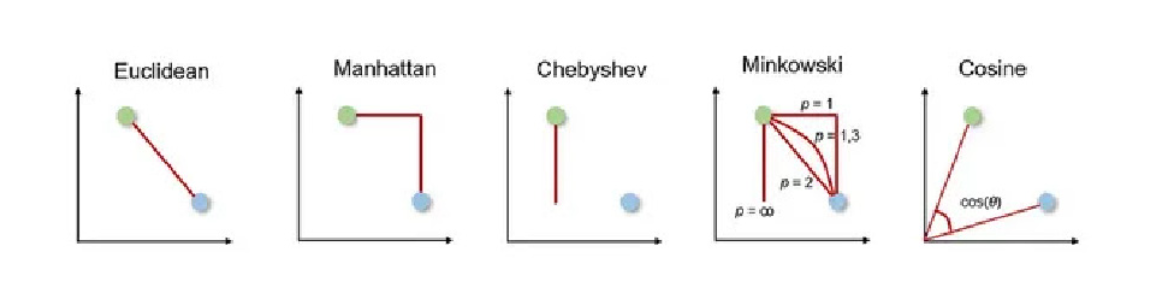
Statistical similarity in data science
Statistical similarity in data science is usually measured using Pearson correlation.
Pearson correlation
Measures the linear relationship between two vectors.

import scipy
scipy.stats.pearsonr((1, 5, 0), (7, 3, 4))(0)
>>> -0.544Other correlation metrics for different types of variables are discussed here.
The metrics mentioned above are effective in measuring the distance between numerical values. However, when it comes to text, we employ different techniques to calculate distance.
To calculate text distance metrics, we can install the necessary libraries using
'pip install textdistance(extras)'
Edit-Based Distance Measurements in Data Science
Now let's look at some edit-based distance measures used in data science.
Hamming distance
Measures the number of different characters between two strings of equal length.
We can add prefixes if we want to calculate strings of unequal length.
textdistance.hamming('series', 'serene')
>>> 3
textdistance.hamming('AGCTTAG', 'ATCTTAG')
>>> 1
textdistance.hamming.normalized_distance('AGCTTAG', 'ATCTTAG')
>>> 0.1428Levenshtein Distance
It is calculated based on how many corrections are needed to convert one string to another. The corrections allowed are insertion, deletion and substitution.
textdistance.levenshtein('genomics', 'genetics')
>>> 2
textdistance.levenshtein('datamining', 'dataanalysis')
>>> 8Damerau-Levenshtein
It also includes the transposition of two adjacent characters in addition to corrections from Levenshtein's distance.
textdistance.levenshtein('algorithm', 'algortihm')
>>> 2
textdistance.damerau_levenshtein('algorithm', 'algortihm')
>>> 1Jaro–Winkler distance
The formula to measure this is Jaro-Winkler=Jaro+(l×p×(1−Jaro)), where
l=common prefix length (up to 4 characters)
p=scale factor, usually 0.1
Jaro = 1/3 (∣s1∣/m + ∣s2∣/m + (m−t)/m), where
If it is the length of the rope
m is the number of matching characters within max(∣s1∣,∣s2∣)/2 – 1
t is the number of transpositions.
For example, in the strings “MARTHA” and “MARHTA”, “T” and “H” are transpositions
textdistance.jaro_winkler('datamining', 'dataanalysis')
>>> 0.6444
textdistance.jaro_winkler('datamining', 'dataanalysis')
>>> 0.8833Token-based distance measures in data science
Let me introduce you to some token-based distance measures in data science.
Jaccard Index
This measures the similarity between two strings by dividing the number of characters common to both by the total number of strings in both (Intersection over Union).
textdistance.jaccard('genomics', 'genetics')
>>> 0.6
textdistance.jaccard('datamining', 'dataanalysis')
>>> 0.375
# The results are similarity fraction between words.Sørensen dice coefficient
measures the similarity between two sets by dividing twice the size of their intersection by the size of their union.
textdistance.sorensen_dice('genomics', 'genetics')
>>> 0.75
textdistance.sorensen_dice('datamining', 'dataanalysis')
>>> 0.5454Tversky index
It is like a generalization of the Sørensen-Dice coefficient and the Jaccard index.
Tversky index(A,B)=∣A∩B∣ / ∣A∩B∣+α∣A−B∣+β∣B−A∣
When alpha and beta are 1, it is the same as the Jaccard index. When they are 0.5 each, it is the same as the Sørensen-Dice coefficient. We can change those values depending on how much weight to give to the discrepancies of A and B, respectively.
textdistance.Tversky(ks=(1,1)).similarity('datamining', 'dataanalysis')
>>> 0.375
textdistance.Tversky(ks=(0.5,0.5)).similarity('datamining', 'dataanalysis')
>>> 0.5454Cosine Similarity
It measures the cosine of the angle between two nonzero vectors in a multidimensional space. cosine_similarity = A⋅B / ∣∣A∣∣×∣∣B∣, AB is the scalar product, ∣∣A∣∣ and ∣∣B∣are the magnitudes.
textdistance.cosine('AGCTTAG', 'ATCTTAG')
>>> 0.8571
textdistance.cosine('datamining', 'dataanalysis')
>>> 0.5477Sequence-based distance measures in data science
We have reached the last section of this article where we will explore some of the commonly used sequence-based distance measures.
Longest common subsequence
This is the longest subsequence common to both strings, where we can get the subsequence by removing zero or more characters without changing the order of the remaining characters.
textdistance.lcsseq('datamining', 'dataanalysis')
>>> 'datani'
textdistance.lcsseq('genomics is study of genome', 'genetics is study of genes')
>>> 'genics is study of gene'Longest common substring
This is the longest substring common to both strings, where we can get a substring in a contiguous sequence of characters within a string.
textdistance.lcsstr('datamining', 'dataanalysis')
>>> 'data'
textdistance.lcsstr('genomics is study of genome', 'genetics is study of genes')
>>> 'ics is study of gen'Ratcliff-Obershelp Similarity
A measure of similarity between two strings based on the concept of matching subsequences. Calculates similarity by finding the longest matching substring between the two strings and then recursively searching for matching substrings in the unmatched segments. The unmatched segments are taken from the left and right parts of the string after dividing the original strings by the matching substring.
Similarity = 2×M / ∣S1∣+∣S2∣
Example:
Chain 1: data mining, Chain 2: data analysis
Longest matching substring: 'data', remaining segments: 'mining' and 'analysis' both on the right side.
Compare mining and analysis, longest matching substring: 'n', remaining segments: 'e' and 'a' on the left side, 'ing' and 'alysis' on the right side. There are no more matching substrings.
So, 2*5 / (10+12) = 0.4545
textdistance.ratcliff_obershelp('datamining', 'dataanalysis')
>>> 0.4545
textdistance.ratcliff_obershelp('genomics is study of genome', 'genetics is study of genes')
>>> 0.8679These are some of the most commonly used similarity and distance metrics in data science. Some others include Smith-Waterman based on dynamic programming, normalized compression distance based on compression, phonetic algorithms like coincidence classification approach, etc.
Learn more about these similarity measures here.
Conclusion
Similarity and dissimilarity measures are crucial in data science for tasks such as clustering and classification. This article explored several metrics: Euclidean and Minkowski distances for numerical data, Pearson correlation for statistical relationships, Hamming and Levenshtein distances for text, and advanced methods such as Jaro-Winkler, Tversky index, and Ratcliff-Obershelp similarity for nuanced comparisons, improving analytical capabilities. .
Frequent questions
A. Euclidean distance is a measure of the straight-line distance between two points in multidimensional space, commonly used in clustering and classification tasks to compare numerical data points.
A. Levenshtein distance measures the number of insertions, deletions, and substitutions required to transform one string into another, while Hamming distance only counts character substitutions and requires that the strings be the same length.
A. The Jaro-Winkler distance measures the similarity between two strings and gives higher scores to strings with matching prefixes. It is particularly useful for comparing names and other text data with common prefixes.
A. Cosine similarity is ideal for comparing document vectors in high-dimensional spaces, such as in information retrieval, text mining, and clustering tasks, where the orientation of the vectors (rather than their magnitude) is important.
A. Token-based similarity measures, such as the Jaccard index and Sørensen-Dice coefficient, compare sets of tokens (words or characters) in strings. They are important for tasks where the presence and frequency of specific elements are crucial, such as in text analysis and document comparison.
 NEWSLETTER
NEWSLETTER




