Today many people have digital access to your medical records, including your doctor’s clinical notes. However, clinical notes are difficult to understand because of the specialized language doctors use, which contains unknown shorthand and abbreviations. In fact, there are thousands of such abbreviations, many of which are specific to certain medical specialties and locations or can mean multiple things in different contexts. For example, a physician might write in his clinical notes, “pt referred to pt by lbp,” which is intended to convey the statement, “Patient referred to physical therapy for low back pain.” Arriving at this translation is difficult for laymen and computers because some abbreviations are uncommon in everyday language (eg, “lbp” stands for “low back pain”), and even familiar abbreviations, such as “pt” for ” patient” may have alternative meanings, such as “physical therapy”. To disambiguate between multiple meanings, the surrounding context must be considered. It is not an easy task to decipher all the meanings, and previous investigations suggests that expanding shorthand and abbreviations may help patients better understand their health, diagnoses, and treatments.
In “Decrypt clinical abbreviations with a privacy-protecting machine learning system“, published in nature communications, we report our findings on a general method that deciphers clinical abbreviations in a way that is both advanced and on par with board-certified physicians in this task. We built the model using only public data on the web that was not associated with any patient (i.e., no potentially sensitive data) and evaluated performance on real, anonymous inpatient and outpatient physician notes from different health systems. To allow the model to generalize from web data to notes, we created a way to algorithmically rewrite large amounts of text from the Internet to look like it was written by a doctor (called reverse substitution at web scale), and we develop a new inference method (called elicitive inference).
 |
| The model input is a string that may or may not contain medical abbreviations. We train a model to generate a corresponding string in which all abbreviations are detected and expanded simultaneously. If the input string does not contain an abbreviation, the model will output the original string. For Rajkomar et al. used under CC BY 4.0/ Cropped from original. |
Rewrite text to include medical abbreviations
Building a system for translating physicians’ notes would typically start with a large data set representative of clinical text where all abbreviations are labeled with their meanings. But there is no such data set for general use by researchers. Therefore, we sought to develop an automated way to create such a data set, but without the use of actual patient notes, which could include sensitive data. We also wanted to ensure that models trained on this data would continue to perform well on actual clinical notes from multiple hospital sites and types of care, such as inpatient and outpatient.
To do this, we referenced a dictionary of thousands of clinical abbreviations and their expansions, and found sentences on the web that contained uses of this dictionary’s expansions. We then “rewrote” those sentences by abbreviating each expansion, resulting in web data that looked like it was written by a doctor. For example, if a website contains the phrase “atrial fibrillation patients may have chest pain,” we would rewrite this sentence as “atrial fibrillation patients may have cp.” We then use the abbreviated text as input to the model, with the original text serving as the label. This approach provided us with large amounts of data to train our model to perform abbreviation expansion.
The idea of ”reversing” long forms with their abbreviations was introduced in previous investigations, but our distributed algorithm allows us to extend the technique to large data sets the size of a web. Our algorithm, called reverse substitution at web scale (WSRS), is designed to ensure that rare terms appear more frequently and common terms are sampled from the public web for a more balanced data set. With this data in hand, we trained a series of large transformer-based language models for expanding web text.
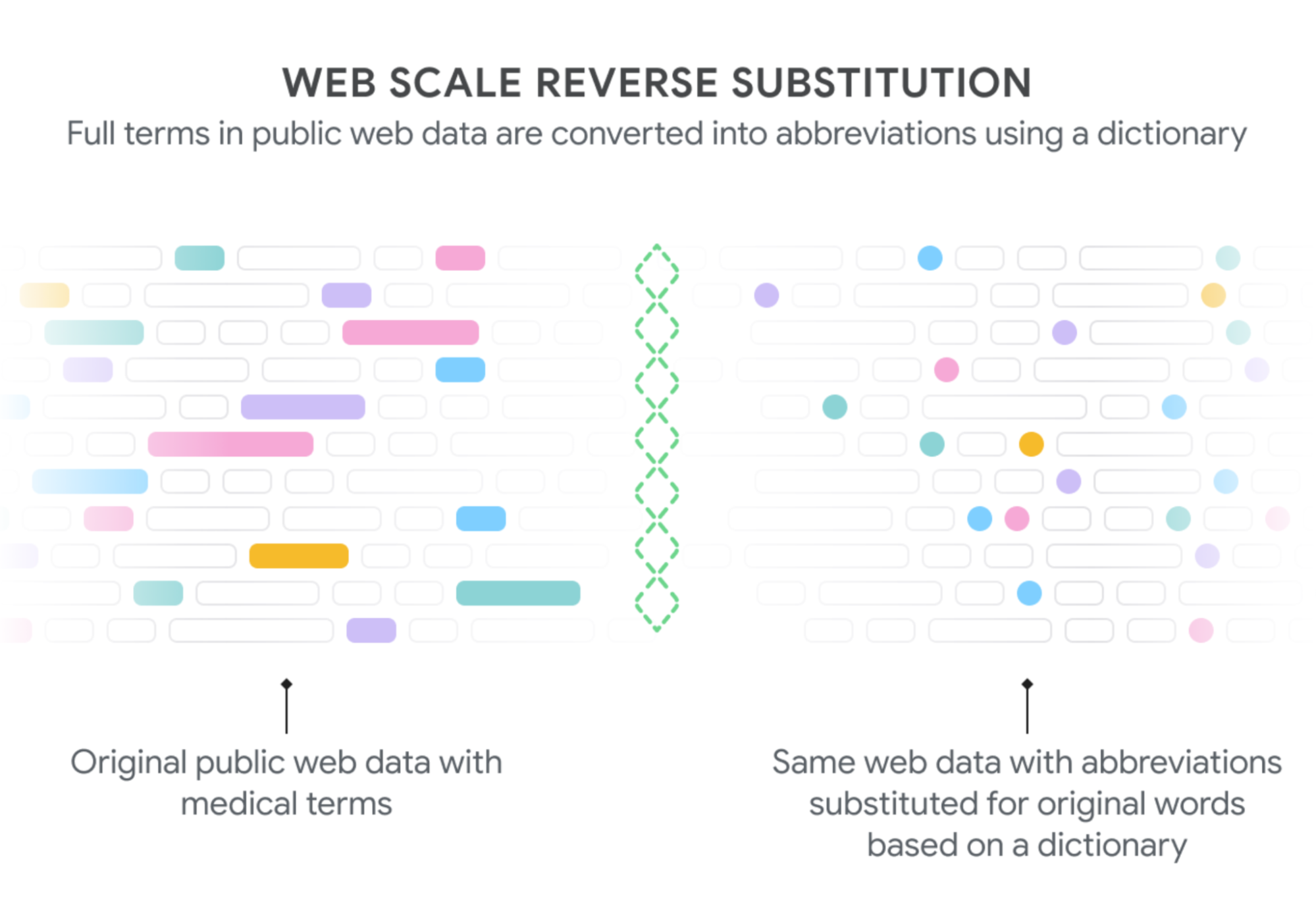 |
| We generate text to train our model on the decoding task by extracting phrases from public web pages that have the corresponding medical abbreviations (shaded boxes on the left) and then replacing them with the appropriate abbreviations (shaded dots, on the right). Since some words are found much more frequently than others (“patient” rather than “posterior tibial”, which can be abbreviated as “pt”), we downsampled the common expansions to obtain a more balanced data set across the thousands of abbreviations. For Rajkomar et al. used under CC BY 4.0. |
Adaptation of protein alignment algorithms to unstructured clinical text
The evaluation of these models in the particular task of abbreviation expansion is difficult. Since they produce unstructured text as output, we had to figure out which abbreviations in the input correspond to which expansion in the output. To accomplish this, we created a modified version of the Needleman’s Desire Algorithmwhich was originally designed for divergent sequence alignment in molecular biology, to align the input and output of the model and extract the corresponding abbreviation-expansion pairs. Using this alignment technique, we were able to assess the model’s ability to accurately detect and expand abbreviations. We evaluated Text-to-Text Transfer Transformer (T5) models of various sizes (ranging from 60 million to over 60 billion parameters) and found that larger models performed better translation than smaller models, with the largest model achieving the best performance.
Creation of new model inference techniques to persuade the model
However, we found something unexpected. When we evaluated performance on multiple sets of external tests from actual clinical notes, we found that the models would leave some abbreviations unexpanded, and for larger models, the problem of incomplete expansion was even worse. This is mainly due to the fact that while we substitute the expansions on the web for their abbreviations, we have no way of handling the abbreviations that are already present. This means that the abbreviations appear in both the original and rewritten text and are used as respective labels and entries, and the model learns not to expand them.
To address this, we developed a new inference chaining technique in which the model’s output is fed back as input to persuade the model to perform further expansions as long as the model is confident of the expansion. In technical terms, our best performing technique, which we call elicitive inferenceinvolves examining the results of a beam search above a certain log-likelihood threshold. Using elicitive inference, we were able to achieve a state-of-the-art ability to expand abbreviations across multiple external test sets.
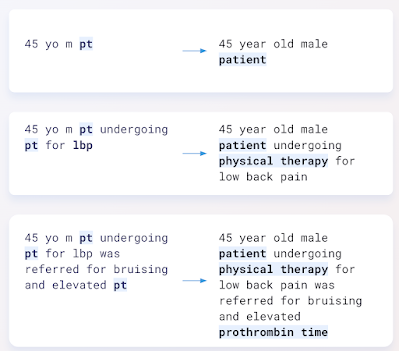 |
| Actual example of model input (left) and output (right). |
comparative performance
We also sought to understand how patients and clinicians currently perform in deciphering clinical notes and how our model compares. We found that lay people (people without specific medical training) demonstrated less than 30% comprehension of the abbreviations present in the sample medical texts. When we let them use Google Search, their comprehension increased to almost 75%, leaving 1 in 5 abbreviations undecipherable. As expected, medical students and trained physicians performed much better on the task with 90% accuracy. We found that our largest model was able to match or beat the experts, with 98% accuracy.
How does the model perform so well compared to the doctors in this task? There are two important factors in the high comparative performance of the model. Part of the discrepancy is that there were some abbreviations that doctors didn’t even try to expand (such as “cm” per centimeter), which partly reduced the measured performance. This may seem unimportant, but for non-English speakers these abbreviations may not be familiar, so it may be helpful to write them down. Rather, our model is designed to comprehensively expand abbreviations. Also, doctors are familiar with the abbreviations they often see in their specialty, but other specialists use abbreviations that are not understood by those outside their fields. Our model is trained on thousands of abbreviations across multiple specialties and can therefore decipher a variety of terms.
Towards better health literacy
We believe there are numerous avenues in which LLMs can help improve patients’ health literacy by increasing the information they see and read. Most LLMs are trained in data that doesn’t look like clinical note data, and the unique distribution of this data makes it difficult to implement these out-of-the-box models. We have shown how to overcome this limitation. Our model also serves to “normalize” clinical note data, enabling additional ML capabilities to make the text easier to understand for patients of all levels of education and health literacy.
Thanks
This work was carried out in collaboration with Yuchen Liu, Jonas Kemp, Benny Li, Ming-Jun Chen, Yi Zhang, Afroz Mohiddin, and Juraj Gottweis. We thank Lisa Williams, Yun Liu, Arelene Chung, and Andrew Dai for many helpful conversations and discussions about this work.
 NEWSLETTER
NEWSLETTER





