Latent diffusion models have increased considerably in popularity in recent years. Due to their outstanding generation capabilities, these models can produce high-fidelity synthetic data sets that can be added to supervised machine learning pipelines in situations where training data is sparse, such as medical imaging. Furthermore, such medical imaging data sets often need to be annotated by trained medical professionals who can decipher small but semantically significant image aspects. Latent diffusion models can provide an easy method to produce synthetic medical imaging data by obtaining relevant medical keywords or concepts of interest.
A Stanford research team investigated the representation limits of large vision and language basic models and evaluated how to use pretrained basic models to represent medical imaging studies and concepts. More specifically, they investigated the representational ability of the stable diffusion model to assess the effectiveness of its language and vision encoders.
The authors used chest radiographs (CXR), the most popular imaging technique worldwide. These CXRs come from two publicly accessible databases, CheXpert and MIMIC-CXR. From each data set, 1000 frontal radiographs with their corresponding reports were randomly selected.

A CLIP text encoder is included with the stable broadcast pipeline (figure above) and parses the text cues to produce a 768-dimensional latent representation. This representation is then used to condition a denoising U-Net to produce images in latent image space using random noise as initialization. Eventually, this latent representation is mapped to pixel space via the decoder component of a variational autoencoder.
First, the authors investigated whether the text encoder is only capable of projecting clinical indications into the latent space of the text while maintaining clinically significant information (1) and whether the VAE is only capable of reconstructing radiological images without losing clinically significant features (2). . Finally, they proposed three techniques to refine the stable diffusion model in the domain of radiology (3).
1 FOOT
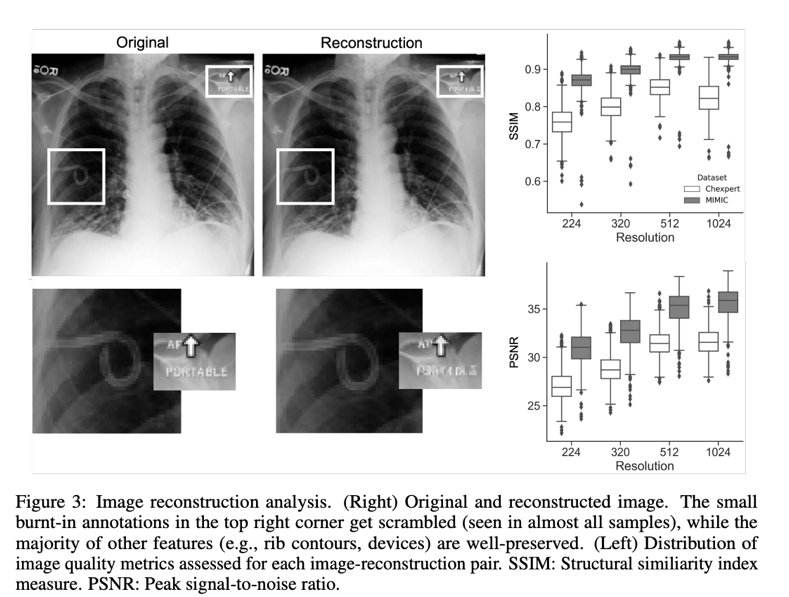
Stable Diffusion, a latent diffusion model, uses a trained encoder to exclude high-frequency details that reflect perceptually insignificant features to transform image inputs into latent space before completing the generative denoising process. Sampled CXR images from CheXpert or MIMIC (“sources”) were coded into latent representations and reconstructed into images (“reconstructions”) to examine how well medical imaging information is preserved while passing through the VAE. The root mean square error (RMSE) and other metrics, such as the Fréchet starting distance (FID), were calculated to objectively measure the quality of the reconstruction, while being qualitatively assessed by a senior radiologist with seven years of experience. A model that had been previously trained to recognize 18 different diseases was used to investigate how the reconstruction procedure affected the classification performance. The following image is an example of a rebuild.
2. Text encoder
The goal of this project is to be able to condition the generation of images on linked medical problems that can be communicated via a text prompt in the specific context of radiology reports and images (eg in report form). Since the rest of the stable diffusion process depends on the ability of the text encoder to accurately represent medical features in latent space, the authors investigated this problem using a technique based on previously trained language models published in the field.
3. Fine adjustment
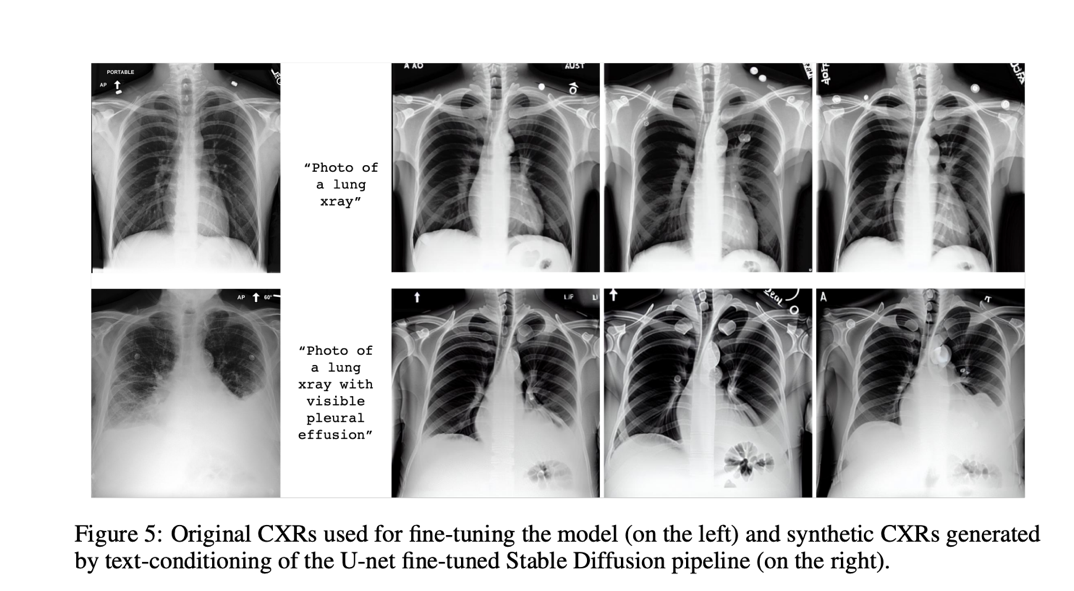
To create domain-specific images, several strategies were tried. In the first experiment, the authors switched the CLIP text encoder, which had been kept frozen during the initial stable diffusion training, with a text encoder that had already been previously trained on data from the biomedical or radiological fields. In the second, text encoder embeds were the main emphasis while tuning the stable broadcast model. In this situation, a new token is introduced that can be used to define features at the patient, procedure, or abnormality level. The third uses domain-specific images to wrap all components besides U-net. After possible fine-tuning by one of the scenarios, the different generative models were tested with two simple prompts: “A photo of a lung X-ray” and “A snapshot of a lung X-ray with a noticeable pleural effusion.” “. .” The models produced synthetic images based solely on this text conditioning. U-Net’s fine-tuning method stands out among the others as the most promising because it achieves the lowest FID scores and, unsurprisingly, produces the most realistic results, demonstrating that such generative models are capable of learning concepts. radiology and can be used to insert realistic-looking abnormalities.
review the Paper. All credit for this research goes to the researchers of this project. Also, don’t forget to join our 17k+ ML SubReddit, discord channeland electronic newsletterwhere we share the latest AI research news, exciting AI projects, and more.
![]()
Leonardo Tanzi is currently a Ph.D. Student at the Polytechnic University of Turin, Italy. His current research focuses on human-machine methodologies for intelligent support during complex interventions in the medical field, using Deep Learning and Augmented Reality for 3D assistance.
 NEWSLETTER
NEWSLETTER





