Image by author
As a data professional, you are probably familiar with the cost of poor data quality. For all data projects, big or small, you need to perform essential data quality checks.
There are dedicated libraries and frameworks for data quality assessment. But if you are a beginner, you can run simple but important data quality checks with pandas. And this tutorial will teach you how to do it.
We will use the California Housing Dataset from scikit-learn for this tutorial.
We will use the California housing dataset from Scikit-learn. data sets module. The data set contains more than 20,000 records of eight numerical characteristics and an objective average home value.
Let’s read the data set into a pandas data frame. df:
from sklearn.datasets import fetch_california_housing
import pandas as pd
# Fetch the California housing dataset
data = fetch_california_housing()
# Convert the dataset to a Pandas DataFrame
df = pd.DataFrame(data.data, columns=data.feature_names)
# Add target column
df('MedHouseVal') = data.targetFor a detailed description of the data set, run data.DESCR as shown:


Data output.DESCR
Let’s get some basic information about the data set:
Here is the result:
Output >>>
RangeIndex: 20640 entries, 0 to 20639
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 MedInc 20640 non-null float64
1 HouseAge 20640 non-null float64
2 AveRooms 20640 non-null float64
3 AveBedrms 20640 non-null float64
4 Population 20640 non-null float64
5 AveOccup 20640 non-null float64
6 Latitude 20640 non-null float64
7 Longitude 20640 non-null float64
8 MedHouseVal 20640 non-null float64
dtypes: float64(9)
memory usage: 1.4 MBBecause we have numerical functions, we will also begin the summary using the describe() method:
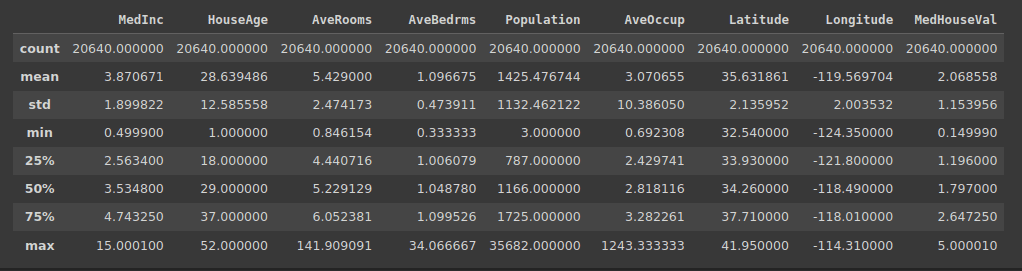

Output of df.describe()
Real-world data sets often have missing values. To analyze the data and build models, it is necessary to handle these missing values.
To ensure data quality, you should check whether the fraction of missing values is within a specific tolerance limit. You can then impute missing values using appropriate imputation strategies.
Therefore, the first step is to check all entities in the data set for missing values.
This code looks for missing values in each column of the data frame. df:
# Check for missing values in the DataFrame
missing_values = df.isnull().sum()
print("Missing Values:")
print(missing_values)The result is a pandas series showing the count of missing values for each column:
Output >>>
Missing Values:
MedInc 0
HouseAge 0
AveRooms 0
AveBedrms 0
Population 0
AveOccup 0
Latitude 0
Longitude 0
MedHouseVal 0
dtype: int64As you can see, there are no missing values in this data set.
Duplicate records in the data set can bias the analysis. Therefore, you should find and discard duplicate records as necessary.
Here is the code to identify and return duplicate rows in df. If there are duplicate rows, they will be included in the result:
# Check for duplicate rows in the DataFrame
duplicate_rows = df(df.duplicated())
print("Duplicate Rows:")
print(duplicate_rows)The result is an empty data frame. Which means there are no duplicate records in the data set:
Output >>>
Duplicate Rows:
Empty DataFrame
Columns: (MedInc, HouseAge, AveRooms, AveBedrms, Population, AveOccup, Latitude, Longitude, MedHouseVal)
Index: ()When analyzing a data set, you will often need to transform or scale one or more entities. To avoid unexpected errors when performing these types of operations, it is important to check if all columns have the expected data type.
This code checks the data types of each column in the data frame. df:
# Check data types of each column in the DataFrame
data_types = df.dtypes
print("Data Types:")
print(data_types)Here, all numerical characteristics are float data type as expected:
Output >>>
Data Types:
MedInc float64
HouseAge float64
AveRooms float64
AveBedrms float64
Population float64
AveOccup float64
Latitude float64
Longitude float64
MedHouseVal float64
dtype: objectOutliers are data points that are significantly different from other points in the data set. If you remember, we ran the describe() method in the data frame.
Based on the quartile values and the maximum value, you could have identified that a subset of features contains outliers. Specifically, these features:
- medinc
- AveRooms
- AveRooms
- Population
One approach to handling outliers is to use the Interquartile rangethe difference between the 75th and 25th quartiles. If Q1 is the 25th quartile and Q3 is the 75th quartile, then the interquartile range is given by: Q3 – Q1.
We then use the quartiles and the IQR to define the interval. (Q1 - 1.5 * IQR, Q3 + 1.5 * IQR). And all points outside this range are outliers.
columns_to_check = ('MedInc', 'AveRooms', 'AveBedrms', 'Population')
# Function to find records with outliers
def find_outliers_pandas(data, column):
Q1 = data(column).quantile(0.25)
Q3 = data(column).quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
outliers = data((data(column) < lower_bound) | (data(column) > upper_bound))
return outliers
# Find records with outliers for each specified column
outliers_dict = {}
for column in columns_to-check:
outliers_dict(column) = find_outliers_pandas(df, column)
# Print the records with outliers for each column
for column, outliers in outliers_dict.items():
print(f"Outliers in '{column}':")
print(outliers)
print("\n")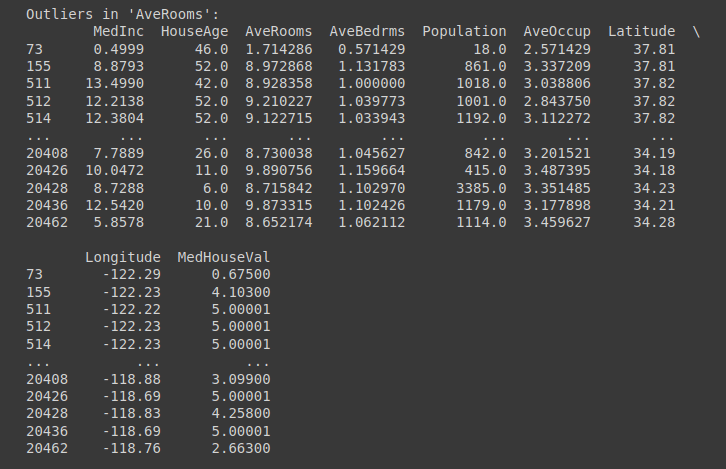

Outliers in ‘AveRooms’ column | Truncated output for outlier checking
An important check of numerical characteristics is to validate the range. This ensures that all observations of a feature take values in an expected range.
This code validates that the ‘MedInc’ value is within an expected range and identifies data points that do not meet this criteria:
# Check numerical value range for the 'MedInc' column
valid_range = (0, 16)
value_range_check = df(~df('MedInc').between(*valid_range))
print("Value Range Check (MedInc):")
print(value_range_check)You can try other numerical functions of your choice. But we see that all the values in the ‘MedInc’ column are in the expected range:
Output >>>
Value Range Check (MedInc):
Empty DataFrame
Columns: (MedInc, HouseAge, AveRooms, AveBedrms, Population, AveOccup, Latitude, Longitude, MedHouseVal)
Index: ()Most data sets contain related features. That’s why it’s important to include checks based on logically relevant relationships between columns (or features).
While features (individually) may take on values in the expected range, the relationship between them may be inconsistent.
Below is an example of our data set. In a valid record, ‘AveRooms’ should normally be greater than or equal to ‘AveBedRms’.
# AveRooms should not be smaller than AveBedrooms
invalid_data = df(df('AveRooms') < df('AveBedrms'))
print("Invalid Records (AveRooms < AveBedrms):")
print(invalid_data)In the California housing data set we are working with, we see that there are no invalid records:
Output >>>
Invalid Records (AveRooms < AveBedrms):
Empty DataFrame
Columns: (MedInc, HouseAge, AveRooms, AveBedrms, Population, AveOccup, Latitude, Longitude, MedHouseVal)
Index: ()Inconsistent data entry is a common data quality problem in most data sets. Examples include:
- Inconsistent formatting in date and time columns
- Inconsistent recording of categorical variable values
- Reading log on different drives.
In our data set, we check the data types of the columns and identify outliers. But you can also perform checks to detect inconsistent data entries.
Let’s prepare a simple example to check if all date entries have a consistent format.
Here we use regular expressions along with pandas. apply() function to check if all date entries are in the YYYY-MM-DD Format:
import pandas as pd
import re
data = {'Date': ('2023-10-29', '2023-11-15', '23-10-2023', '2023/10/29', '2023-10-30')}
df = pd.DataFrame(data)
# Define the expected date format
date_format_pattern = r'^\dtechnology-\deducation tech-\deducation tech$' # YYYY-MM-DD format
# Function to check if a date value matches the expected format
def check_date_format(date_str, date_format_pattern):
return re.match(date_format_pattern, date_str) is not None
# Apply the format check to the 'Date' column
date_format_check = df('Date').apply(lambda x: check_date_format(x, date_format_pattern))
# Identify and retrieve entries that do not follow the expected format
non_adherent_dates = df(~date_format_check)
if not non_adherent_dates.empty:
print("Entries that do not follow the expected format:")
print(non_adherent_dates)
else:
print("All dates are in the expected format.")This returns entries that do not follow the expected format:
Output >>>
Entries that do not follow the expected format:
Date
2 23-10-2023
3 2023/10/29In this tutorial, we go over common data quality checks with Pandas.
When working on smaller data analysis projects, these data quality checks with pandas are a good starting point. Depending on the problem and data set, it may include additional checks.
If you are interested in learning about data analysis, check out the guide 7 Steps to Master Data Management with Pandas and Python.
Bala Priya C. is a developer and technical writer from India. He enjoys working at the intersection of mathematics, programming, data science, and content creation. His areas of interest and expertise include DevOps, data science, and natural language processing. He likes to read, write, code and drink coffee! Currently, he is working to learn and share his knowledge with the developer community by creating tutorials, how-to guides, opinion pieces, and more.
 NEWSLETTER
NEWSLETTER