
Image by the author
Imagine you're trying to solve a puzzle that's missing pieces. It can be frustrating, right? It's a common scenario when working with incomplete datasets. Masked arrays in NumPy are specialized array structures that allow you to handle missing or invalid data efficiently. They're especially useful in situations where you need to perform calculations on datasets that contain unreliable inputs.
A masked array is essentially a combination of two arrays:
- Data matrix:The main array containing the actual data values.
- Mask matrix:A boolean array of the same shape as the data array, where each element indicates whether the corresponding data item is valid or masked (invalid/missing).
Data matrix
The data array is the main component of a masked array, which contains the actual data values that you want to analyze or manipulate. This array can contain any numerical or categorical data, just like a standard NumPy array. Here are some important points to keep in mind:
- Storage:The data array stores the values you need to work with, including valid and invalid entries (such as “NaN” or specific values that represent missing data).
- OperationsWhen performing operations, NumPy uses the data array to calculate the results, but will consider the mask array to determine which elements to include or exclude.
- Compatibility:The data array in a masked array supports all standard NumPy functionality, making it easy to switch between regular and masked arrays without significantly altering your existing codebase.
Example:
import numpy as np
data = np.array((1.0, 2.0, np.nan, 4.0, 5.0))
masked_array = np.ma.array(data)
print(masked_array.data) # Output: ( 1. 2. nan 4. 5.)
Mask matrix
The mask matrix is a boolean matrix of the same form as the data matrix. Each element of the mask matrix corresponds to an element of the data matrix and indicates whether that element is valid (FAKE) or masked (TRUE) Some points are detailed below:
- Structure:The mask matrix is created in the same shape as the data matrix to ensure that each data point has a corresponding mask value.
- Indicating invalid data: TO TRUE The value in the mask array marks the corresponding data point as invalid or missing, while a FAKE The value indicates valid data. This allows NumPy to ignore or exclude invalid data points during calculations.
- Automatic masking:NumPy provides functions to automatically create mask arrays based on specific conditions (e.g.,
np.ma.masked_invalid()to mask Yaya values).
Example:
import numpy as np
data = np.array((1.0, 2.0, np.nan, 4.0, 5.0))
mask = np.isnan(data) # Create a mask where NaN values are True
masked_array = np.ma.array(data, mask=mask)
print(masked_array.mask) # Output: (False False True False False)
The power of masked arrays lies in the relationship between the data and the masked arrays. When performing operations on a masked array, NumPy considers both arrays to ensure that calculations are based only on valid data.
Benefits of masked matrices
Masked arrays in NumPy offer several advantages, especially when working with datasets containing missing or invalid data, some of which include:
- Efficient handling of missing data:Masked arrays allow you to easily flag invalid or missing data, such as NaNs, and automatically handle them in calculations. Operations are performed only on valid data, ensuring that missing or invalid entries do not skew the results.
- Data cleansing made easy:Functions like
numpy.ma.masked_invalid()You can automatically mask common invalid values (e.g. NaN or infinity) without requiring additional code to manually identify and handle these values. You can define custom masks based on specific criteria, allowing for flexible data cleansing strategies. - Seamless integration with NumPy functions:Masked arrays work with most standard NumPy functions and operations. This means you can use familiar NumPy methods without manually excluding or preprocessing masked values.
- Greater precision in calculations:When performing calculations (e.g. mean, sum, standard deviation), masked values are automatically excluded from the calculation, resulting in more accurate and meaningful results.
- Improved data visualization:When visualizing data, masked matrices ensure that invalid or missing values are not graphed, resulting in clearer and more accurate visual representations. You can graph only valid data, avoiding clutter and improving the interpretation of graphs and charts.
Using masked arrays to handle missing data in NumPy
In this section we will demonstrate how to use a masked array to handle missing data in Numpy. First, let's look at a simple example:
import numpy as np
# Data with some missing values represented by -999
data = np.array((10, 20, -999, 30, -999, 40))
# Create a mask where -999 is considered as missing data
mask = (data == -999)
# Create a masked array using the data and mask
masked_array = np.ma.array(data, mask=mask)
# Calculate the mean, ignoring masked values
mean_value = masked_array.mean()
print(mean_value)
Production:
25.0
Explanation:
- Data creation:
datais an array of integers where -999 Represents missing values. - Creating masks:
maskis a boolean array that marks positions with -999 as TRUE (indicating missing data). - Creating a masked array:
np.ma.array(data, mask=mask)creates a masked array, applying the mask todata. - Calculation:
masked_array.mean().
calculates the mean ignoring masked values (i.e., -999), resulting in the average of the remaining valid values.
In this example, the mean is calculated only from (10, 20, 30, 40)Excluding -999 values.
Let's explore a more complete example where masked matrices are used to handle missing data in a larger dataset. We'll use a scenario involving a dataset of temperature readings from multiple sensors over several days. The dataset contains some missing values due to sensor failures.
Use case: Analysis of temperature data from multiple sensors
Script:You have temperature readings from five sensors over ten days. Some readings are missing due to sensor issues. We need to calculate the average daily temperature by ignoring the missing data.
Data set:The dataset is represented as a 2D NumPy array, with rows representing days and columns representing sensors. Missing values are indicated by np.nan.
Steps to follow:
- Import NumPy:For matrix operations and handling masked matrices.
- Defining the data:Create a 2D array of temperature readings with some missing values.
- Create a mask:Identify missing (NaN) values in the dataset.
- Create masked matrices:Apply mask to handle missing values.
- Calculate Daily Averages Calculates the average temperature for each day, ignoring missing values.
- Output results:Displays the results for analysis.
Code:
import numpy as np
# Example temperature readings from 5 sensors over 10 days
# Rows: days, Columns: sensors
temperature_data = np.array((
(22.1, 21.5, np.nan, 23.0, 22.8), # Day 1
(20.3, np.nan, 22.0, 21.8, 23.1), # Day 2
(np.nan, 23.2, 21.7, 22.5, 22.0), # Day 3
(21.8, 22.0, np.nan, 21.5, np.nan), # Day 4
(22.5, 22.1, 21.9, 22.8, 23.0), # Day 5
(np.nan, 21.5, 22.0, np.nan, 22.7), # Day 6
(22.0, 22.5, 23.0, np.nan, 22.9), # Day 7
(21.7, np.nan, 22.3, 22.1, 21.8), # Day 8
(22.4, 21.9, np.nan, 22.6, 22.2), # Day 9
(23.0, 22.5, 21.8, np.nan, 22.0) # Day 10
))
# Create a mask for missing values (NaNs)
mask = np.isnan(temperature_data)
# Create a masked array
masked_data = np.ma.masked_array(temperature_data, mask=mask)
# Calculate the average temperature for each day, ignoring missing values
daily_averages = masked_data.mean(axis=1) # Axis 1 represents days
# Print the results
for day, avg_temp in enumerate(daily_averages, start=1):
print(f"Day {day}: Average Temperature = {avg_temp:.2f} °C")
Production:
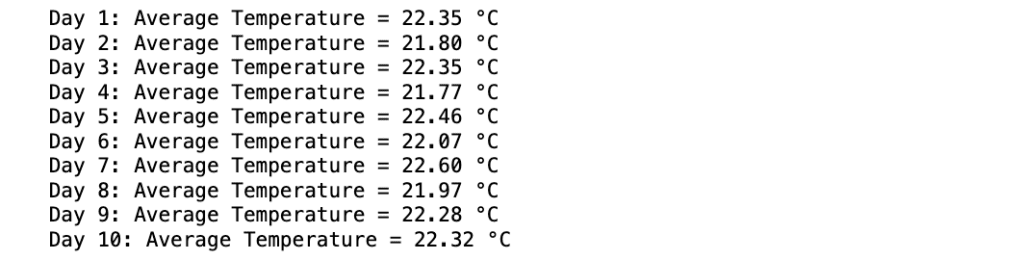

Explanation:
- Import NumPy:Import the NumPy library to use its functions.
- Define data:Create a 2D array
temperature_datawhere each row represents the temperatures of the sensors on a specific day and some values are missing (np.nan). - Create mask:Generate a boolean mask using
np.isnan(temperature_data)to identify missing values (TRUE where are the valuesnp.nan). - Create a masked array: Wear
np.ma.masked_array(temperature_data, mask=mask)createmasked_dataThis array masks missing values, allowing operations to ignore them. - Calculate daily averages:Calculate the average temperature of each day using
.mean(axis=1). Here,axis=1means calculating the average of the sensors for each day. - Output results: Print the average temperature for each day. Masked values are excluded from the calculation, providing accurate daily averages.
Conclusion
In this article, we explore the concept of masked matrices and how they can be leveraged to deal with missing data. We discuss the two key components of masked matrices: the data matrix, which contains the actual values, and the mask matrix, which indicates which values are valid or missing. We also examine their benefits, including efficient handling of missing data, seamless integration with NumPy functions, and improved computational accuracy.
We demonstrate the use of masked matrices through simple and more complex examples. The initial example illustrated how to handle missing values represented by specific markers such as -999while the most complete example showed how to analyze temperature data from multiple sensors, where missing values are indicated by np.nanBoth examples highlighted the ability of masked matrices to accurately calculate results by ignoring invalid data.
For more information, please see these two resources:
Olumida of Shittu Shittu is a software engineer and technical writer passionate about leveraging cutting-edge technologies to create compelling narratives, with a keen eye for detail and a knack for simplifying complex concepts. You can also find Shittu on twitter.com/Shittu_Olumide_”>twitter.
<script async src="//platform.twitter.com/widgets.js” charset=”utf-8″>






