Genomic research is a fundamental field that focuses on understanding the structure, function and evolution of genomes. It encompasses studies on DNA sequences, genetic variations, and the intricate mechanisms that govern gene expression and regulation. This field has profound implications for biotechnology, medicine, and evolutionary biology, offering insights into genetic disorders, potential therapies, and fundamental life processes.
A critical issue is the need for advanced models to predict and generate biological sequences. Current methods can be more complex and scalable to accurately model genomic functions. Researchers are seeking solutions to improve the accuracy and efficiency of these models to better understand and manipulate biological systems.
Current methods often need more capacity to handle the complexity and scale needed to accurately model genomic functions. Researchers are seeking solutions to improve the accuracy and efficiency of these models to better understand and manipulate biological systems. Traditional approaches in genomic modeling have primarily used modality-specific models focused on proteins, regulatory DNA, or RNA. These models often need help handling multi-scale interactions in complex biological processes. Generative applications have been restricted to the design of simple molecules and short sequences, lacking the breadth necessary for complete genomic analysis.
Researchers from Stanford University, the Arc Institute, TogetherAI, CZ Biohub, and the University of California, Berkeley, have presented evoa genomic-based model designed to perform prediction and generation tasks from the molecular to genomic scale. evo leverages a novel deep signal processing architecture to handle vast genomic data sets with high precision. evoThe architecture incorporates a hybrid of attention mechanisms and convolutional operators, allowing it to process sequences at single nucleotide resolution in long contexts. Trained on 7 billion parameters with data from entire prokaryotic genomes, evo It can generalize across DNA, RNA, and protein modalities, allowing it to predict genetic functions and generate complex biological systems.
evo It employs a state-of-the-art deep signal processing architecture, StripeHyena, which combines attention mechanisms with convolutional operators to process long genomic sequences efficiently. This hybrid approach allows evo to maintain high resolution at the single nucleotide level, which is crucial for capturing detailed variations in genetic sequences. The model is based on extensive prokaryotic genome data sets totaling 300 billion nucleotide tokens, including bacterial and archaeal genomes and millions of predicted phage and plasmid sequences. This comprehensive training allows evo learn the intricate patterns of genomic sequences, making it capable of predicting and generating tasks in different molecular modalities. The training process involved two stages: initially a context length of 8,000 tokens was used and extended to 131,000 tokens to capture broader genomic contexts. evoThe architecture of includes 29 layers of data-driven convolutional operators interleaved with multi-head attention layers equipped with rotating position embeddings, improving its ability to remember long sequence information.
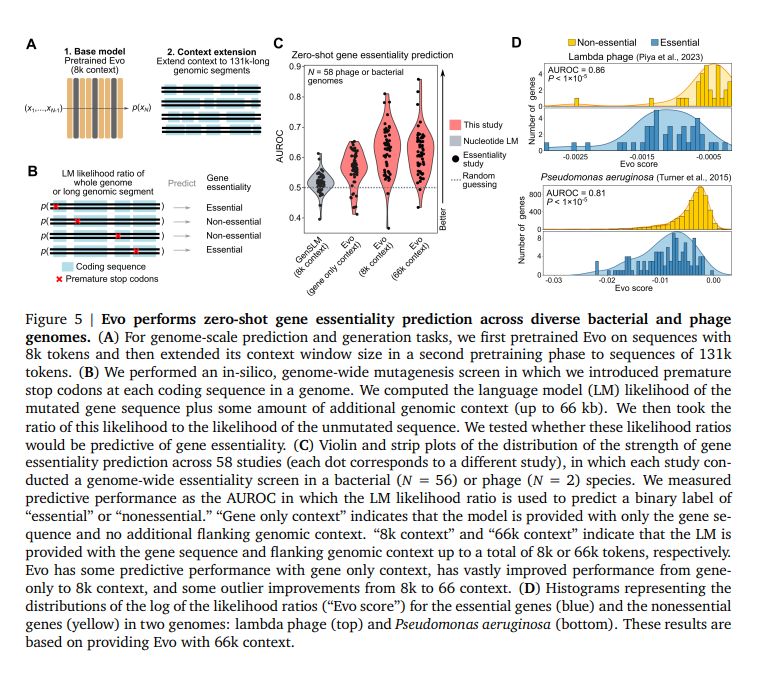
The performance of evo excels in zero-shot function generation and prediction tasks. It can generate synthetic CRISPR-Cas molecular complexes and transposable systems, predict genetic essentiality with high precision, and create coding-rich sequences up to 650 kilobases in length. In terms of specific performance metrics, evo demonstrated a Spearman correlation of 0.64 in predicting the fitness effects of 5S ribosomal RNA mutations in E. coli. For the prediction of gene expression, evo achieved a correlation of 0.41 for mRNA expression and an AUROC of 0.68 for protein expression prediction. The model's ability to predict genetic essentiality was also impressive, with an AUROC of 0.86 for lambda phage essentiality and 0.81 for Pseudomonas aeruginosa. These capabilities exceed those of existing domain-specific language models, highlighting evoThe advanced performance of on various genomic tasks. Besides, evoThe generative capabilities of are demonstrated by their ability to produce coherent CRISPR-Cas systems, with 15-45% of the generated sequences containing Cas coding sequences of up to 5 kb and generating transposable elements with significant protein sequence diversity. .
In conclusion, the research team has developed a powerful tool in evo which addresses the limitations of previous models. By enabling comprehensive genomic analysis and generation, evo represents a significant advance in the field and promises to improve our understanding and control of biological systems at multiple levels. evoThe success of Modeling Genomic Data at Scale and its ability to perform zero predictions and generate complex biological sequences marks a major advance in genomics research. This model not only provides a deeper mechanistic understanding of biology, but also accelerates the potential for engineering life forms, offering a new paradigm in biological research and synthetic biology.
Review the Paper. All credit for this research goes to the researchers of this project. Also, don't forget to follow us on twitter.com/Marktechpost”>twitter. Join our Telegram channel, Discord Channeland LinkedIn Grabove.
If you like our work, you will love our Newsletter..
Don't forget to join our 42k+ ML SubReddit
![]()
Nikhil is an internal consultant at Marktechpost. He is pursuing an integrated double degree in Materials at the Indian Institute of technology Kharagpur. Nikhil is an ai/ML enthusiast who is always researching applications in fields like biomaterials and biomedical science. With a strong background in materials science, he is exploring new advances and creating opportunities to contribute.
<script async src="//platform.twitter.com/widgets.js” charset=”utf-8″>
 NEWSLETTER
NEWSLETTER





