When a patient is diagnosed with cancer, one of the most important steps is examination of the tumor under a microscope by pathologists to determine cancer. scenery and characterize the tumor. This information is critical to understanding the clinical prognosis (ie, the likely patient outcomes) and to determining the most appropriate treatment, such as surgery alone versus surgery plus chemotherapy. The development of machine learning (ML) tools in pathology to assist with microscopic review represents a compelling research area with many potential applications.
Previous studies have shown that ML can pinpoint identify and classify tumors on pathology images and can even predict patients forecast wearing acquaintance pathological features, such as degree to which the appearance of the gland deviates from normal. While these efforts are focused on using ML to detect or quantify known features, alternative approaches offer the potential to identify novel characteristics. The discovery of new features could, in turn, further improve cancer prognosis and treatment decisions for patients by extracting information not yet considered in current workflows.
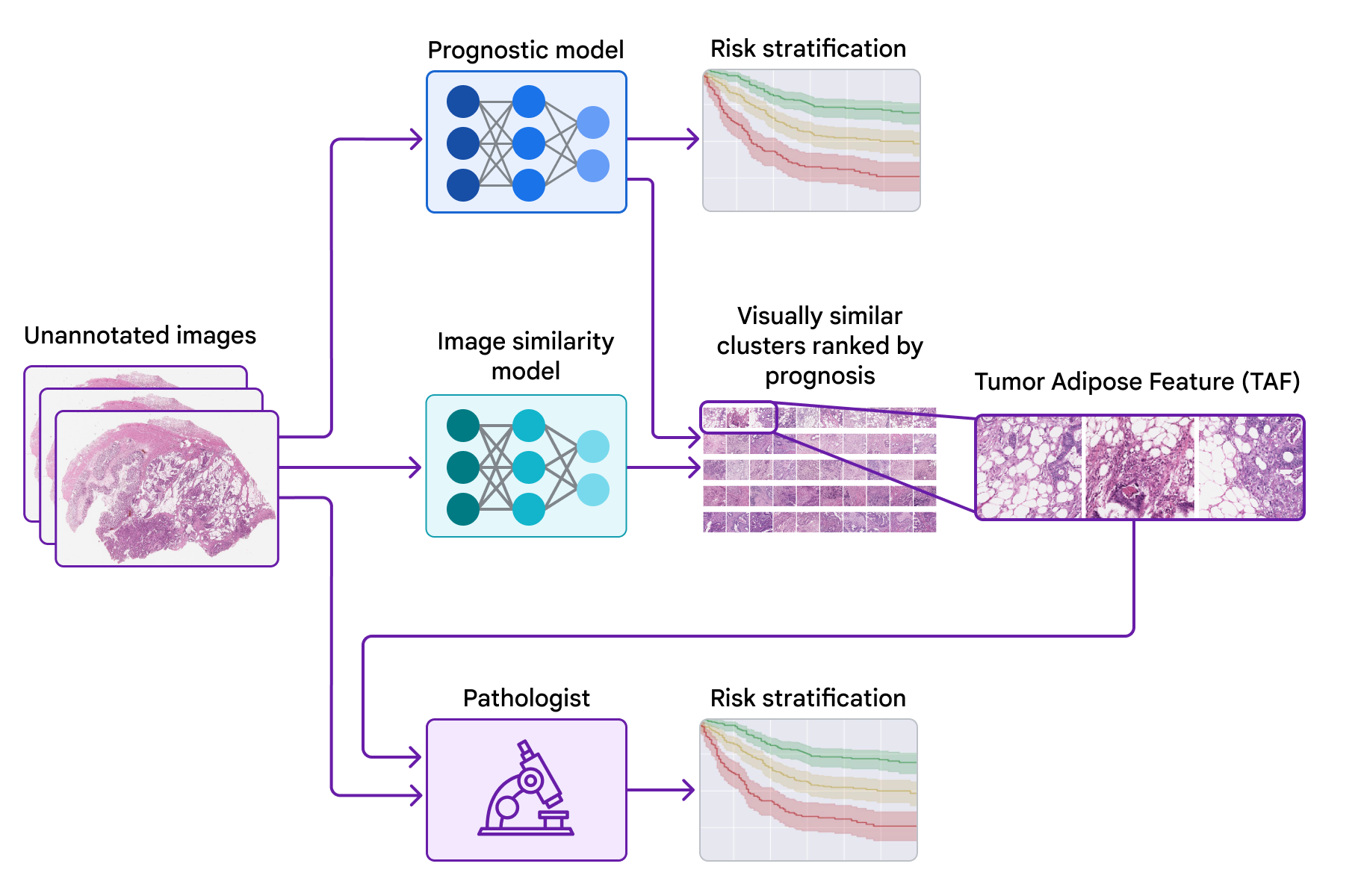
Today, we would like to share the progress we have made in recent years in identifying novel features for colorectal cancer in collaboration with teams from the Graz Medical University in Austria and the University of Milano-Bicocca (UNIMIB) in Italy. Next, we will cover several stages of the work: (1) train a model to predict prognosis from pathology images without specifying the features to use, so that it can learn which features are important; (2) test that forecast model using explainability techniques; and (3) identify a new feature and validate its association with the patient’s prognosis. We describe this feature and evaluate its use by pathologists in our recently published article, “Pathological validation of a machine learning function for colon cancer risk stratification”. To our knowledge, this is the first demonstration that medical experts can learn new prognostic functions from machine learning, a promising start to the future of this “learn from deep learning” paradigm.
Training a forecast model to know which features are important
One potential approach to identify novel features is to train ML models to directly predict patient outcomes using only the images and paired outcome data. This is in contrast to training models to predict human-annotated “intermediate” labels for acquaintance pathologic features and then use those features to predict outcomes.
Initial work by our team showed the feasibility of training models for directly predict the prognosis of a variety of cancers using the publicly available TCGA data set. It was especially exciting to see that, for some types of cancer, the model’s predictions were prognostic after controlling for available pathologic and clinical features. Together with collaborators of the Graz Medical University and the Biobank Grazlater we expanded this work using a large anonymous colorectal cancer cluster. Interpreting these model predictions became an intriguing but common next step. interpretability techniques they were difficult to apply in this context and did not provide clear insights.
Interpretation of the learned characteristics of the model
To test the features used by the forecast model, we use a second model (trained to identify image similarity) to cluster patches of large pathology images. We then used the forecast model to calculate the ML-predicted average risk score for each group.
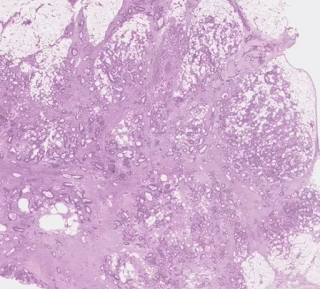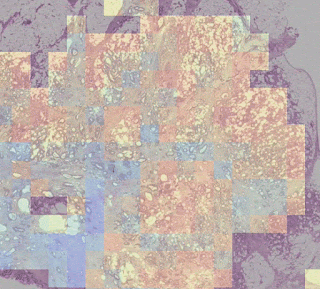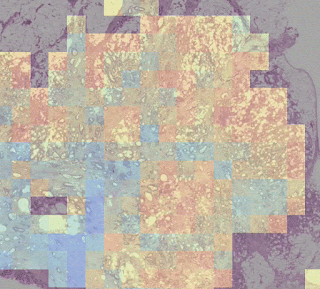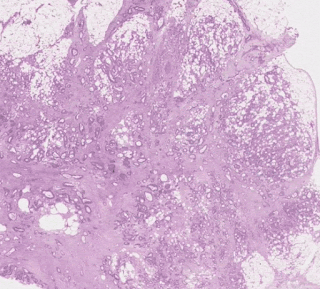
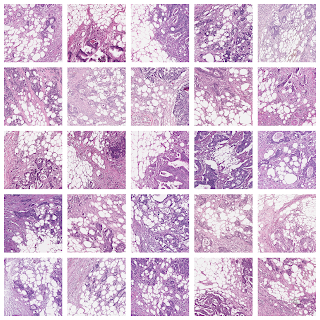
One group stood out for its high average risk score (associated with a poor prognosis) and distinctive visual appearance. The pathologists described the images as involving a high-grade tumor (ie, normal tissue that looked less like it) in close proximity to adipose (fat) tissue, leading us to call this group “tumor adipose feature” (TAF). ); see the following figure for detailed examples of this feature. Further analysis showed that the relative amount of TAF was itself a strong and independent predictor.
| Left: H&E pathology slide with a heat map overlaid indicating tumor adipose feature (TAF) locations. The image similarity model considers regions highlighted in red/orange to be more likely to have TAF, compared to regions highlighted in green/blue or regions that are not highlighted at all. Good: Representative collection of TAF patches in various cases. |
Validation that pathologists can use the learned function of the model
These studies provided a compelling example of the potential of ML models to predict patient outcomes and a methodological approach to gain insight into model predictions. However, intriguing questions remained as to whether pathologists could learn and score the feature identified by the model while maintaining demonstrable prognostic value.
In our most recent articleWe collaborate with pathologists from the NIMIB to investigate these questions. Using TAF example images from the previous post To learn and understand this feature of interest, UNIMIB pathologists developed scoring guidelines for TAF. If TAF was not seen, the case was scored ‘absent’, and if TAF was seen, the categories ‘unifocal’, ‘multifocal’, and ‘generalized’ were used to indicate the relative amount. Our study showed that pathologists could reproducibly identify ML-derived TAF and that its score for TAF provided statistically significant prognostic value in an independent retrospective data set. To our knowledge, this is the first demonstration of pathologists learning to identify and score a specific pathology feature originally identified by an ML-based approach.
Putting things in context: learning from deep learning as a paradigm
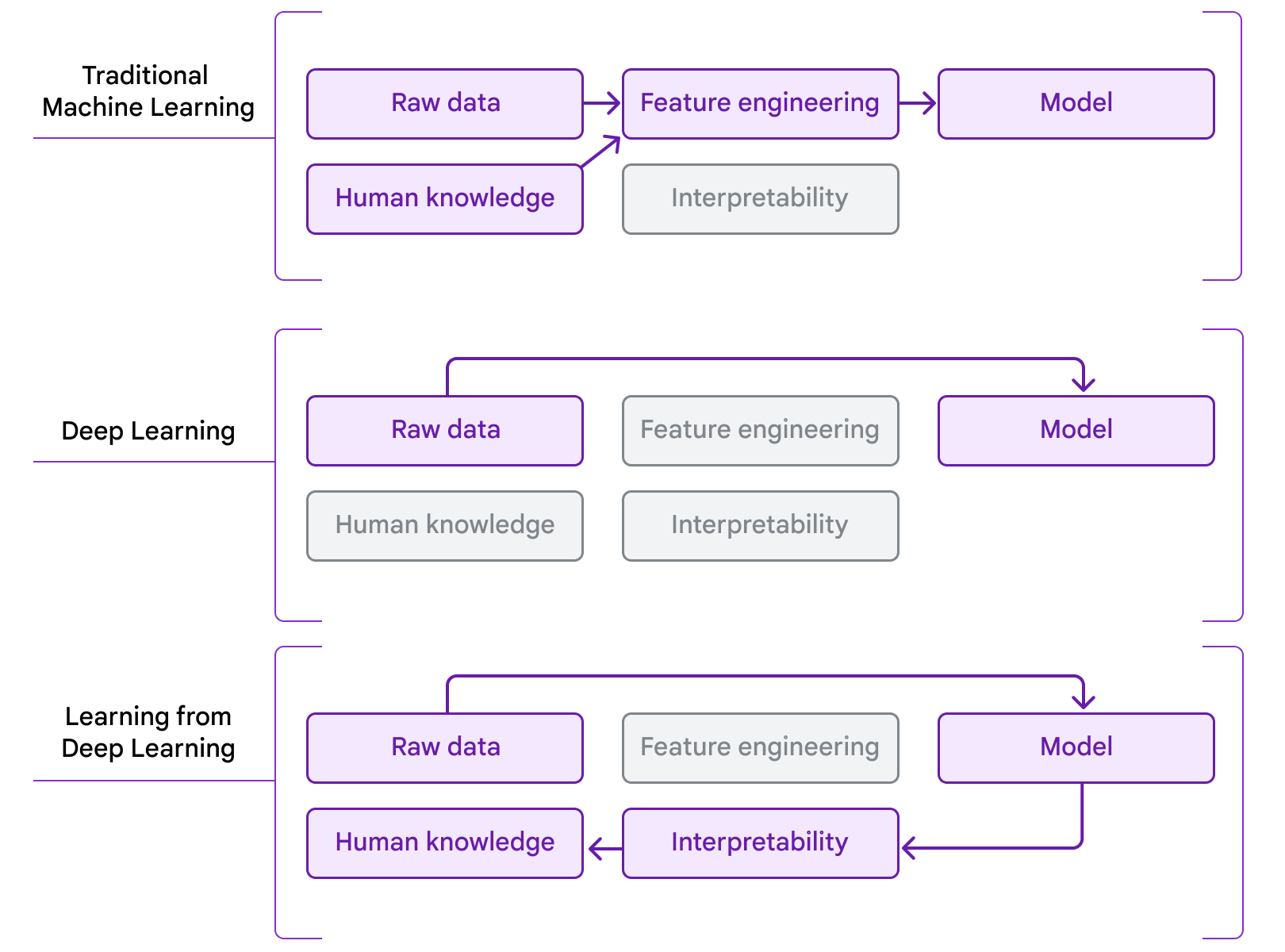
Our work is an example of people who “learn from deep learning”. In traditional machine learning, models learn from hand-drawn features informed by existing domain knowledge. More recently, in the era of deep learning, a combination of large-scale model architectures, computation, and datasets has made it possible to learn directly from raw data, but this is often at the expense of human interpretability. Our work combines the use of deep learning to predict patient outcomes with interpretability methods, to extract new insights that pathologists could apply. We see this process as a natural next step in the evolution of the application of ML to problems in medicine and science, moving from using ML to distill existing human knowledge to people using ML as a tool for knowledge discovery.
Thanks
This work would not have been possible without the efforts of co-authors Vincenzo L’Imperio, Markus Plass, Heimo Muller, Nicolò’ Tamini, Luca Gianotti, Nicola Zucchini, Robert Reihs, Greg S. Corrado, Dale R. Webster, Lily H. Peng , Po-Hsuan Cameron Chen, Marialuisa Lavitrano, David F. Steiner, Kurt Zatloukal, Fabio Pagni. We also appreciate the support of the Verily Life Sciences and Google Health Pathology teams, particularly Timo Kohlberger, Yunnan Cai, Hongwu Wang, Kunal Nagpal, Craig Mermel, Trissia Brown, Isabelle Flament-Auvigne, and Angela Lin. We are also grateful for comments on the manuscript from Akinori Mitani, Rory Sayres, and Michael Howell, and illustration assistance from Abi Jones. This work would also not have been possible without the support of Christian Guelly, Andreas Holzinger, Robert Reihs, Farah Nader, the Biobank Graz, the efforts of the Medical University Graz slide digitization team, the participation of the pathologists who reviewed and annotated cases during the development of the model, and the technicians of the UNIMIB team.