The GraphRAG Python package by Neo4j offers a comprehensive solution for building end-to-end workflows, from transforming unstructured data into a knowledge graph to enabling knowledge graph retrieval and implementing complete GraphRAG pipelines. Whether you’re developing knowledge assistants, search APIs, chatbots, or report generators in Python, this package simplifies the integration of knowledge graphs to enhance the relevance, accuracy, and explainability of retrieval-augmented generation (RAG).
In this guide, we’ll demonstrate how to get started with the GraphRAG Python package, build a GraphRAG pipeline from scratch, and explore various knowledge graph retrieval methods to customize the behavior of your GenAI application.

GraphRAG: Enhancing GenAI with Knowledge Graphs
By combining knowledge graphs with RAG, GraphRAG addresses common challenges of large language models (LLMs), such as hallucinations, while enriching responses with domain-specific context for better quality and precision than traditional RAG methods. Knowledge graphs provide essential contextual data, enabling LLMs to deliver reliable answers and act as trusted agents in complex tasks. Unlike conventional RAG solutions that focus on fragmented textual data, GraphRAG integrates both structured and semi-structured data into the retrieval process.
With the GraphRAG Python package, you can create knowledge graphs and implement advanced retrieval methods, including graph traversals, query generation via text-to-Cypher, vector searches, and full-text searches. The package also includes tools for building complete RAG pipelines, enabling seamless integration of GraphRAG with Neo4j into GenAI workflows and applications.
Key Components of the GraphRAG Knowledge Graph Construction Pipeline
The GraphRAG knowledge graph (KG) construction pipeline consists of several components, each vital in transforming raw text into structured data for enhanced Retrieval-Augmented Generation (RAG)- GraphRAG with Neo4j. These components work together to enable advanced retrieval methods like graph-based searches and context-aware responses. Below are the core components:
- Document Parser: Extracts text from various document formats (e.g., PDFs).
- Document Chunker: Splits the text into smaller pieces that fit within the LLM’s token limit.
- Chunk Embedder (Optional): Computes vector embeddings for each chunk, enabling semantic matching.
- Schema Builder: Defines the structure of the KG, grounding entity extraction and ensuring consistency.
- LexicalGraphBuilder (Optional): Builds a lexical graph connecting documents and chunks.
- Entity and Relation Extractor: Identifies entities (e.g., people, dates) and their relationships.
- Knowledge Graph Writer: Saves the entities and relations to the graph database for retrieval.
- Entity Resolver: Merges duplicate or similar entities into a single node to maintain graph integrity.
Entity Resolver: Merges duplicate or similar entities into a single node to maintain graph integrity.
These components work together to create a dynamic knowledge graph that powers GraphRAG, enabling more accurate and context-aware responses from LLMs.
Set Up a Neo4j Database
To begin the RAG workflow, the first step is to set up a database for retrieval. Neo4j AuraDB provides an easy way to launch a free Graph Database. Depending on the requirements, one can opt for AuraDB Free for basic use or try AuraDB Professional (Pro), which offers increased memory and better performance for ingestion and retrieval tasks. While the Pro version is ideal for optimal results due to its advanced features, for this project, I will utilize Neo4j AuraDB’s free Graph Database.It is a fully managed cloud service that offers a scalable and high-performance graph database solution. With its free tier, users can easily build and explore graph-based applications, leveraging powerful relationships between data points for insights and analysis.
Upon logging into Neo4j AuraDB, you can create a free instance. Once the instance is set up, you’ll receive or can download the necessary credentials, including the username, Neo4j URL, and password, to connect to your database.
Install the Required Libraries
We will install several libraries using pip, including Neo4j’s Python Driver and OpenAI to create GraphRAG with Neo4j & Python. This is an essential step for setting up our environment.
!pip install fsspec openai numpy torch neo4j-graphrag
Set Up Connection Details for Neo4j
NEO4J_URI = ""
username = ""
password = ""
In this section, we have to define the connection details for Neo4j. Replace the placeholders with your actual Neo4j database credentials:
- NEO4J_URI: URI to your Neo4j instance (e.g., bolt://localhost:7687).
- username and password: Your Neo4j authentication credentials.
Set OpenAI API Key
import os
os.environ('OPENAI_API_KEY') = ''
Here, we’re loading OpenAI API key using os.environ. This allows us to use OpenAI’s models for entity extraction in your knowledge graph.
1. Building and Defining the Knowledge Graph Pipeline
To facilitate our research on the greenhouse effect to show GraphRAG with Neo4j & Python, we will transform research papers into a structured knowledge graph and store it in a Neo4j database. Using a selection of PDF documents focused on greenhouse effect studies; we’ll organize the domain-specific data these documents contain into a graph that enhances ai-driven applications. This approach allows for better structuring and retrieval of complex scientific information.
The knowledge graph will include key node types:
- Document: Captures metadata related to the document sources.
- Chunk: Represents text segments from the documents, embedded with vector representations for efficient retrieval.
- Entity: Extracted entities from the text chunks, providing structured context and connections.
To automate the creation of this knowledge graph, we define a SimpleKGPipeline class. This class enables seamless knowledge graph construction by requiring a few essential inputs:
- A Neo4j driver to connect to the Neo4j database.
- An LLM (Language Model) for entity extraction.
- An embedding model to convert text into vectors, enabling similarity searches.
By combining the document transformation with an automated pipeline, we can build a comprehensive knowledge graph that efficiently organizes and retrieves insights about the greenhouse effect.
Neo4j Driver Initialization
import neo4j
from neo4j_graphrag.llm import OpenAILLM
from neo4j_graphrag.embeddings.openai import OpenAIEmbeddings
driver = neo4j.GraphDatabase.driver(NEO4J_URI, auth=(username, password))
Here, we initialize the Neo4j database driver using the NEO4J_URI, username, and password set earlier. We can also import components needed for LLM-based entity extraction (OpenAILLM) and embedding (OpenAIEmbeddings).
Initialize LLM and Embedding Model
llm = OpenAILLM(
model_name="gpt-4o-mini",
model_params={"response_format": {"type": "json_object"}, "temperature": 0},
)
embedder = OpenAIEmbeddings()
We have initialized the LLM (OpenAILLM) for entity extraction and set parameters like the model name (GPT-4o-mini) and response format. The embedder is initialized with OpenAIEmbeddings, which will be used to convert text chunks into vectors for similarity search.
Setting Node Labels
Let’s define different categories of nodes based on our use case:
basic_node_labels = ("Object", "Entity", "Group", "Person", "Organization", "Place")
academic_node_labels = ("ArticleOrPaper", "PublicationOrJournal")
climate_change_node_labels = ("GreenhouseGas", "TemperatureRise", "ClimateModel", "CarbonFootprint", "EnergySource")
node_labels = basic_node_labels + academic_node_labels + climate_change_node_labels
Here, we’ve grouped our node labels into:
- Basic node labels: Generic entity types such as “Person”, “Organization”, etc.
- Academic node labels: Related to academic publications like articles or journals.
- Climate change node labels: Specific to climate change-related entities.
These labels will help categorize entities within your knowledge graph.
Defining Relationship Types
rel_types = ("AFFECTS", "CAUSES", "ASSOCIATED_WITH", "DESCRIBES", "PREDICTS", "IMPACTS")We have defined possible relationships between nodes in the graph. These relationships describe how entities interact or are connected.
Creating the Prompt Template
prompt_template=""'
You are a climate researcher tasked with extracting information from research papers and structuring it in a property graph.
Extract the entities (nodes) and specify their type from the following text.
Also extract the relationships between these nodes.
Return the result as JSON using the following format:
{{"nodes": ( {{"id": "0", "label": "entity type", "properties": {{"name": "entity name"}} }} ),
"relationships": ({{"type": "RELATIONSHIP_TYPE", "start_node_id": "0", "end_node_id": "1", "properties": {{"details": "Relationship details"}} }}) }}
Input text:
{text}
'''
Here, we defined a prompt template for the LLM. The model will be given a text (research paper), and it needs to extract:
- Entities (nodes): These are identified by type (e.g., Person, Organization) and their properties (e.g., name).
- Relationships: The LLM will identify how the entities are related (e.g., “CAUSES”, “ASSOCIATED_WITH”).
Create the Knowledge Graph Pipeline
from neo4j_graphrag.experimental.components.text_splitters.fixed_size_splitter import FixedSizeSplitter
from neo4j_graphrag.experimental.pipeline.kg_builder import SimpleKGPipeline
Here, we’re importing the necessary classes:
- FixedSizeSplitter: This will help split large text (from PDFs) into smaller chunks.
- SimpleKGPipeline: This is the main class for building your knowledge graph.
Building the Knowledge Graph Pipeline
kg_builder_pdf = SimpleKGPipeline(
llm=llm,
driver=driver,
text_splitter=FixedSizeSplitter(chunk_size=500, chunk_overlap=100),
embedder=embedder,
entities=node_labels,
relations=rel_types,
prompt_template=prompt_template,
from_pdf=True
)
- llm: Language model used for entity extraction (you already initialized it with OpenAI’s LLM).
- driver: The Neo4j driver that connects to your Neo4j instance.
- text_splitter: You use FixedSizeSplitter to break down large text from the PDFs into chunks of 500 tokens with an overlap of 100 tokens.
- embedder: Embedding model used to convert the text chunks into vector embeddings.
- entities: Specifies the node labels that define the entities in your knowledge graph.
- relations: Specifies the relationship types that connect the nodes in the graph.
- prompt_template: The template for instructing the LLM to extract nodes and relationships.
- from_pdf=True: Tells the pipeline to extract data from PDF files.
Processing PDFs
In this, we are using three different research papers on Greenhouse:
pdf_file_paths = ('/home/janvi/Downloads/ToxipediaGreenhouseEffectArchive.pdf',
'/home/janvi/Downloads/3.1.pdf',
'/home/janvi/Downloads/Shell_Climate_1988.pdf')
for path in pdf_file_paths:
print(f"Processing: {path}")
pdf_result = await kg_builder_pdf.run_async(file_path=path)
print(f"Result: {pdf_result}")

This loop processes the three PDF files and feeds them into the SimpleKGPipeline. It uses run_async to process the documents asynchronously and prints the result for each document.
Once complete, you can explore the resulting knowledge graph. The Unified Console provides a great interface for this.
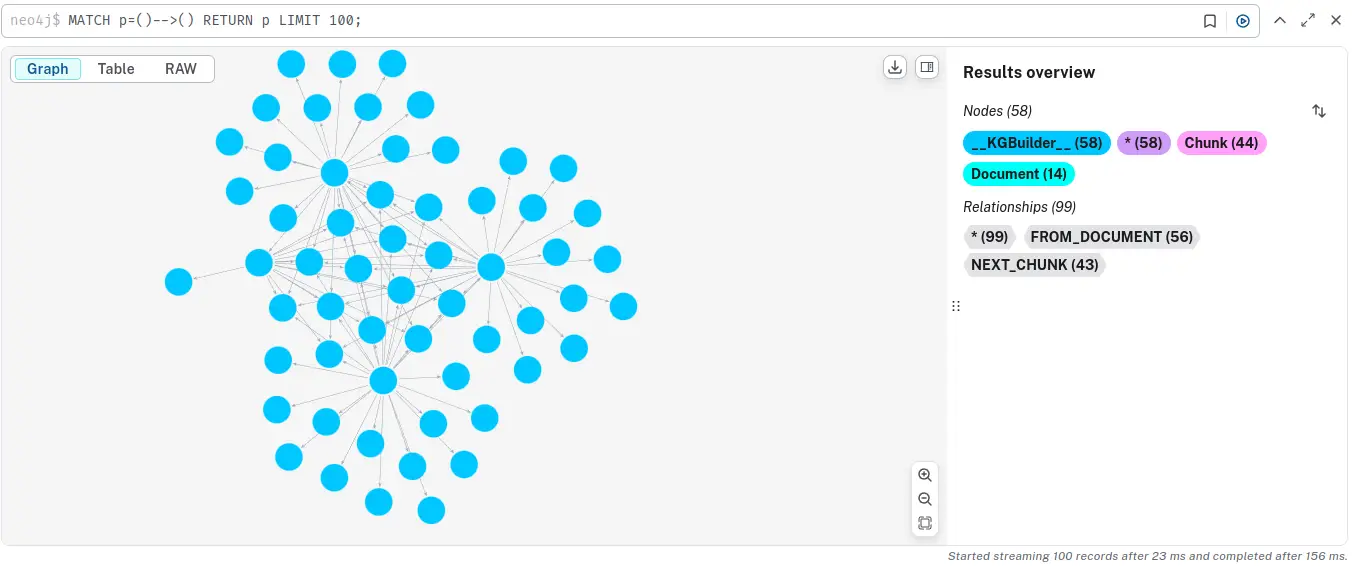
Go to the Query tab and enter the below query to see a sample of the graph.
MATCH p=()-->() RETURN p LIMIT 100;
You can see how the Document, Chunk, and __Entity__ nodes are all connected together.
To see the “lexical” portion of the graph containing Document and Chunk nodes, run the following.
MATCH p=(:Chunk)--(:!__Entity__) RETURN p;
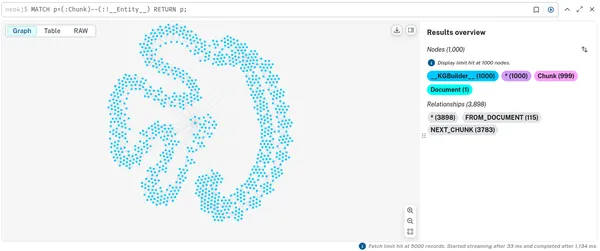
Note that these are disconnected components, one for each document we ingested. You can also see the embeddings that have been added to all chunks.
To look at just the domain graph of __Entity__ nodes, you can run the following:
MATCH p=(:!Chunk)-->(:!Chunk) RETURN p;
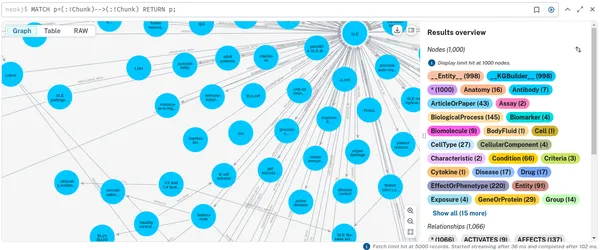
You will see how different concepts have been extracted and how they connect to one another. This domain graph connects information between the documents.
2. Retrieving Data From Your Knowledge Graph
Once the knowledge graph for greenhouse effect research is built, the next step involves retrieving meaningful information to support analysis. The GraphRAG Python package provides versatile retrieval mechanisms tailored to your needs. These include:
- Vector Retriever: Conducts similarity searches using vector embeddings for efficient data retrieval.
- Vector Cypher Retriever: Combines vector search with Cypher queries, Neo4j’s graph query language, enabling graph traversal to include related nodes and relationships in the retrieval.
- Hybrid Retriever: Merges vector and full-text search for comprehensive data retrieval.
- Hybrid Cypher Retriever: Combines hybrid search with Cypher queries for advanced graph traversal.
- Text2Cypher: Converts natural language queries into Cypher queries, enabling users to retrieve data directly from Neo4j without manual query writing.
- Weaviate & Pinecone Neo4j Retriever: Integrates vector searches from external systems like Weaviate or Pinecone with Neo4j nodes using external ID properties.
- Custom Retriever: Offers flexibility for implementing tailored retrieval methods for specific needs.
These retrieval mechanisms empower the implementation of diverse retrieval patterns, improving the relevance and accuracy of retrieval-augmented generation (RAG) pipelines.
Vector Retriever and Knowledge Graph Retrieval
For our greenhouse effect research knowledge graph, we utilize the Vector Retriever, which uses Approximate Nearest Neighbor (ANN) vector search. This retriever retrieves data by performing similarity searches on embeddings associated with text chunks stored in the graph.
Setting Up a Vector Index
To enable vector-based retrieval, we create a Vector Index in Neo4j. This index operates on the text chunks in the graph, allowing the Vector Retriever to pull back relevant insights with high precision.
By combining Neo4j’s vector search capabilities and these retrieval methods, we can query the knowledge graph to extract valuable information about the causes, effects, and solutions related to the greenhouse effect.
from neo4j_graphrag.indexes import create_vector_index
create_vector_index(driver, name="text_embeddings", label="Chunk",
embedding_property="embedding", dimensions=1536, similarity_fn="cosine")
create_vector_index: This function creates a vector index on the Chunk label in Neo4j. The embeddings (generated from the PDF text) will be stored in the embedding property of each Chunk node. The index is based on cosine similarity, and the embeddings have a dimension of 1536, which is standard for OpenAI’s embeddings.
Using the VectorRetriever
from neo4j_graphrag.retrievers import VectorRetriever
vector_retriever = VectorRetriever(
driver,
index_name="text_embeddings",
embedder=embedder,
return_properties=("text"),
)
VectorRetriever: This component queries the Chunk nodes using vector search, which allows us to find the most relevant chunks based on the input query. The return_properties parameter ensures that the search results will return the text of the chunk.
Searching for Information in the Knowledge Graph
import json
vector_res = vector_retriever.get_search_results(
query_text="What are the main greenhouse gases contributing to the Greenhouse Effect and their impacts as discussed in the documents?",
top_k=3
)
for i in vector_res.records:
print("====\n" + json.dumps(i.data(), indent=4))
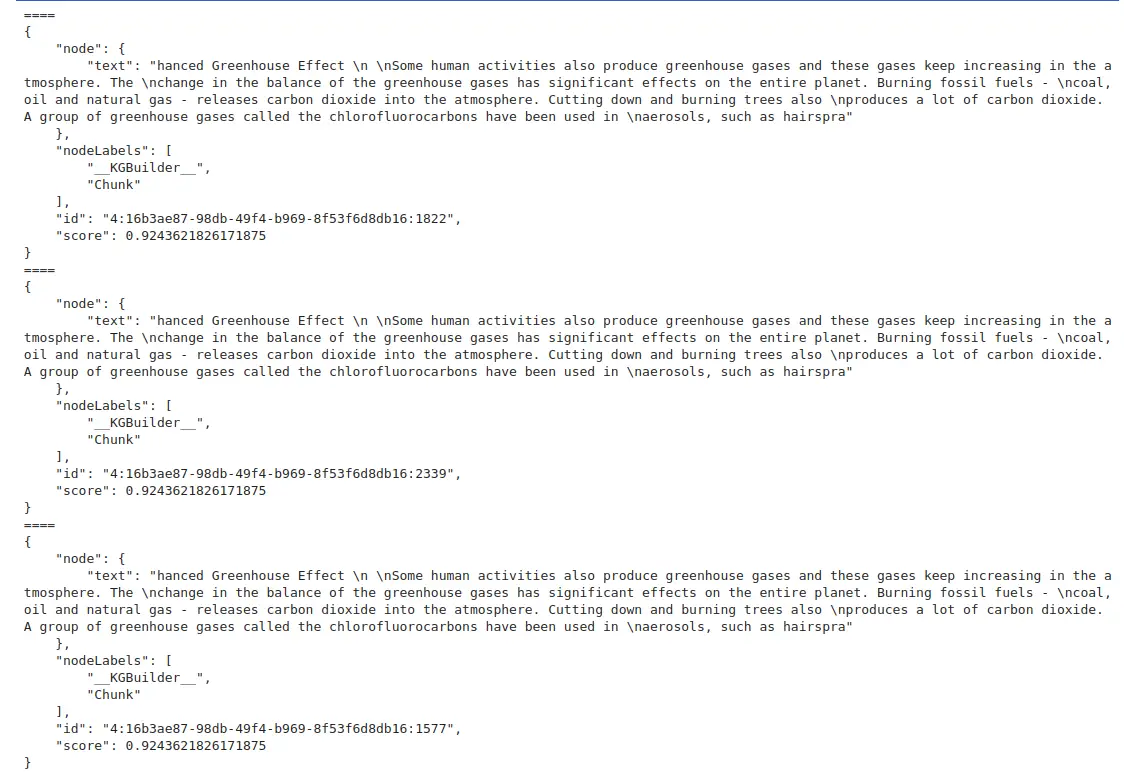
- get_search_results: This function performs a vector search with the input query (in this case, asking about greenhouse gases and their impacts).
- top_k=3: We are limiting the number of results to the top 3 most relevant chunks.
- The results are printed in a nicely formatted JSON structure, which includes the relevant text and metadata of the retrieved chunks.
Using the VectorCypherRetriever for Graph Traversal
The VectorCypherRetriever allows for an advanced method of knowledge graph retrieval by combining vector search with Cypher queries. This enables us to traverse the graph based on semantic similarities found in the text, exploring related entities and their relationships.
Setting up the VectorCypherRetriever
from neo4j_graphrag.retrievers import VectorCypherRetriever
vc_retriever = VectorCypherRetriever(
driver,
index_name="text_embeddings",
embedder=embedder,
retrieval_query="""
// 1) Go out 2-3 hops in the entity graph and get relationships
WITH node AS chunk
MATCH (chunk)<-(:FROM_CHUNK)-()-(relList:!FROM_CHUNK)-{1,2}()
UNWIND relList AS rel
// 2) Collect relationships and text chunks
WITH collect(DISTINCT chunk) AS chunks,
collect(DISTINCT rel) AS rels
// 3) Format and return context
RETURN '=== text ===\n' + apoc.text.join((c in chunks | c.text), '\n---\n') + '\n\n=== kg_rels ===\n' +
apoc.text.join((r in rels | startNode(r).name + ' - ' + type(r) + '(' + coalesce(r.details, '') + ')' + ' -> ' + endNode(r).name ), '\n---\n') AS info
"""
)
- retrieval_query: This Cypher query is used to define the logic of traversing the graph. Here, you traverse 2-3 hops away from each chunk and capture the relationships between the chunks.
- Text and Relationship Formatting: The results are formatted to return the chunk text first, followed by the relationships encountered during the traversal.
Running a Query for Relevant Information
vc_res = vc_retriever.get_search_results(
query_text="What are the causes and consequences of the Greenhouse Effect as discussed in the provided documents?",
top_k=3
)
- get_search_results: This method performs a vector search based on the input query. It will return the top 3 most relevant chunks and their associated relationships in the knowledge graph.
Extracting and Printing Results
kg_rel_pos = vc_res.records(0)('info').find('\n\n=== kg_rels ===\n')
# Print the results, separating the text chunk context and the KG context
print("# Text Chunk Context:")
print(vc_res.records(0)('info')(:kg_rel_pos))
print("# KG Context From Relationships:")
print(vc_res.records(0)('info')(kg_rel_pos:))
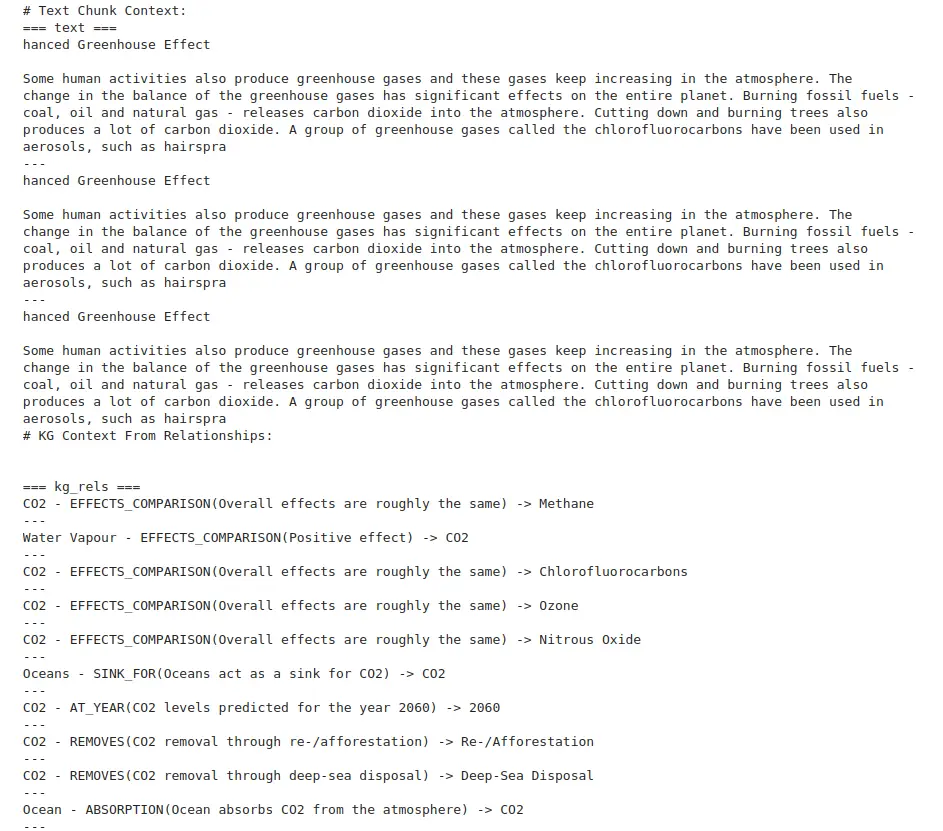
- kg_rel_pos: This locates where the relationships start in the response.
- The results are then printed, separating the textual context from the relationships found in the knowledge graph.
3. Constructing a GraphRAG Pipeline
To further enhance the retrieval-augmented generation (RAG) process for our greenhouse effect research, we now integrate both the VectorRetriever and VectorCypherRetriever into a GraphRAG pipeline. This integration allows us to retrieve relevant data and use that context to generate responses that are strictly based on the knowledge graph, ensuring accuracy and reliability in the generated answers.
Instantiating and Running GraphRAG
The GraphRAG Python package simplifies the process of instantiating and running RAG pipelines. You can easily create a GraphRAG pipeline by utilizing the GraphRAG class. At its core, the class requires two essential components:
- LLM (Language Model): This is responsible for generating natural language responses based on the retrieved context.
- Retriever: This is used to fetch relevant information from the knowledge graph (e.g., using VectorRetriever or VectorCypherRetriever).
Setting up the GraphRAG Pipeline
from neo4j_graphrag.llm import OpenAILLM as LLM
from neo4j_graphrag.generation import RagTemplate
from neo4j_graphrag.generation.graphrag import GraphRAG
llm = LLM(model_name="gpt-4o", model_params={"temperature": 0.0})
rag_template = RagTemplate(template=""'Answer the Question using the following Context. Only respond with information mentioned in the Context. Do not inject any speculative information not mentioned.
# Question:
{query_text}
# Context:
{context}
# Answer:
''', expected_inputs=('query_text', 'context'))
- RagTemplate: The template ensures that the LLM only responds based on the provided context, avoiding any speculative answers.
- GraphRAG: The GraphRAG class uses a language model and a retriever to pull in context to answer the query. It is initialized with both a vector_retriever and vc_retriever.
Creating the GraphRAG Pipelines
v_rag = GraphRAG(llm=llm, retriever=vector_retriever, prompt_template=rag_template)
vc_rag = GraphRAG(llm=llm, retriever=vc_retriever, prompt_template=rag_template)
- v_rag: Uses the VectorRetriever to search for relevant text chunks and answer questions.
- vc_rag: Uses the VectorCypherRetriever to both search for relevant text and traverse relationships in the knowledge graph.
Now we will be executing queries using both the VectorRetriever and VectorCypherRetriever through the GraphRAG pipeline to retrieve context and generate answers from the knowledge graph. Here’s a breakdown of the code:
Query 1: “List the causes, effects, and solutions for the Greenhouse Effect.”This query checks the answers provided by both the vector-based retrieval and vector + Cypher graph traversal methods:
q = "List the causes, effects, and solutions for the Greenhouse Effect."
print(f"Vector Response: \n{v_rag.search(q, retriever_config={'top_k':5}).answer}")
print("\n===========================\n")
print(f"Vector + Cypher Response: \n{vc_rag.search(q, retriever_config={'top_k':5}).answer}")

Query 2: “Explain the Greenhouse Effect in detail. Include its natural process, human-induced causes, global warming impacts, and climate change effects as discussed in the provided documents.”Here, we’re asking for a more detailed explanation. The return_context=True flag is used to return the context along with the answer:
q = "Explain the Greenhouse Effect in detail. Include its natural process, human-induced causes, impacts on global warming, and its effects on climate change as discussed in the provided documents."
v_rag_result = v_rag.search(q, retriever_config={'top_k': 5}, return_context=True)
vc_rag_result = vc_rag.search(q, retriever_config={'top_k': 5}, return_context=True)
print(f"Vector Response: \n{v_rag_result.answer}")
print("\n===========================\n")
print(f"Vector + Cypher Response: \n{vc_rag_result.answer}")

Exploring Retrieved Content: After getting the context results, we’re printing and parsing the contents from the vector and Cypher retrievers:
for i in v_rag_result.retriever_result.items:
print(json.dumps(eval(i.content), indent=1))

For the vc_rag_result, we’re splitting the content and filtering for any text containing the keyword “treat”:
vc_ls = vc_rag_result.retriever_result.items(0).content.split('\\n---\\n')
for i in vc_ls:
if "treat" in i:
print(i)

Query 3: “Can you summarize the Greenhouse Effect?”Finally, we’re summarizing the information requested by the user in list format. Similar to previous queries, we’re retrieving the results and printing the answers:
q = "Can you summarize the Greenhouse Effect? Include its natural process, greenhouse gases involved, impacts on the environment and human health, and challenges in addressing climate change. Provide in list format with details for each item."
print(f"Vector Response: \n{v_rag.search(q, retriever_config={'top_k': 5}).answer}")
print("\n===========================\n")
print(f"Vector + Cypher Response: \n{vc_rag.search(q, retriever_config={'top_k': 5}).answer}")

Conclusion
This article explored how the GraphRAG Python package (GraphRAG with Neo4j) can effectively enhance the retrieval-augmented generation (RAG) process by integrating knowledge graphs with large language models (LLMs). We demonstrated how to create a knowledge graph from research documents related to the Greenhouse Effect and how to store and manage this graph using Neo4j(GraphRAG with Neo4j). By defining the knowledge graph pipeline and leveraging various retrieval methods, such as VectorRetriever and VectorCypherRetriever, we showed how to retrieve relevant information from the graph to generate accurate and contextually relevant responses.
Combining knowledge graphs with RAG helps address common issues such as hallucinations and provides domain-specific context that improves the quality of responses. Additionally, by incorporating multiple retrieval techniques, we enhanced the accuracy and relevance of the generated content, making it more reliable and useful for answering complex questions related to the greenhouse effect.
Overall, GraphRAG with Neo4j offers a powerful toolset for building knowledge-powered applications that require both accurate data retrieval and natural language generation. Incorporating Neo4j’s graph capabilities ensures that responses are contextually grounded and informed by structured and semi-structured data, offering a more robust solution than traditional RAG methods.
Frequently Asked Questions
Ans. GraphRAG is a Python package combining knowledge graphs with retrieval-augmented generation (RAG) to enhance the accuracy and relevance of responses to large language models (LLMs). It retrieves relevant information from knowledge graphs, processes it, and uses it to provide contextually grounded answers to queries. This combination helps mitigate issues like hallucinations, which are common in traditional LLM-based solutions.
Ans. Neo4j is a powerful graph database that efficiently stores and manages relationships between entities, making it an ideal platform for creating knowledge graphs. It supports advanced graph queries using Cypher, which allows for powerful data retrieval and graph traversal. GraphRAG with Neo4j allows you to leverage its capabilities to integrate both structured and semi-structured data into your RAG workflows.
Ans. GraphRAG offers several retrievers for various data retrieval patterns:
Vector Retriever
Vector Cypher Retriever
Hybrid Retriever
Hybrid Cypher Retriever
Text2Cypher
Custom Retriever
Ans. GraphRAG addresses the issue of hallucinations by providing LLMs with structured, domain-specific data from knowledge graphs. Instead of relying solely on the language model’s internal knowledge, GraphRAG ensures that the model generates responses based on reliable and relevant information stored in the graph. This makes the responses more accurate and contextually grounded.
Ans. The Hybrid Retriever combines vector search and full-text search to retrieve data more comprehensively. This method allows GraphRAG to pull both vector-based similar data and traditional textual information, improving the retrieval process’s accuracy and depth. It’s particularly useful when dealing with complex queries requiring diverse context data sources.







